Mining Chromatographic Enantioseparation Data Using Matched Molecular Pair Analysis
Abstract
:1. Introduction
2. Results and Discussion
3. Experimental Section
3.1. Source of Data
3.2. Definition of Enantioselectivity
3.3. Selection of Data to Model
3.4. Transformation of Data
3.5. Method for Matched Molecular Pairs
- Pairs of molecules A and B are selected such that A and B are topologically similar (similarity > 0.65 using the atom-pair descriptor and the Dice similarity index). This step is useful because finding maximum common substructure (MCS) is slow and the selection reduces the number of pairs that must be examined by MCS.
- A MCS is defined for each pair A-B. The change in structure A → B is defined by the RECS (remainder after elimination of common substructure), which is the set of atoms that remain in A and B after the atoms in the MCS are eliminated. One can think of A → B as the “change in structure”. Some A → B changes may be discarded if the changes occur in more than one place on the molecule.
- Sets of A → B pairs are clustered such that all pairs in each cluster have the same type of chemical change, all the pairs are distinct, and all the molecules in the cluster are topologically similar. That is, each pair in the cluster contains the same change in the same molecular context. Here we will use Npair to be the number of pairs in a cluster. Some clusters may contain a single pair. Nclusters will represent the number of clusters from a particular dataset.So far, the analysis has ignored activity. Further steps take the activity into account.
- Each molecule in the cluster is assigned an activity. Consider a cluster of three pairs A1 → B1, A2 → B2, A3 → B3. One can calculate the difference in activity between A1 and B1, A2 and B2, etc. The mean difference in activity for the cluster is called meanDiff and the standard deviation is called StdDiff. If the absolute value of meanDiff is large and StdDiff is small, the same chemical change is producing an equivalent change in activity. A metric Z compares meanDiff of the cluster with Npair pairs with the meanDiff expected by assembling Npair randomly selected pairs not necessarily in the same cluster:Z = (meanDiff(real) − meanDiff(random))/StdDiff(random,Npairs)For randomly selected pairs, the changes in activity will tend to cancel out, so the expected meanDiff(random) is close to zero and StdDiff will have a value that depends on Npairs. As a consequence, as Npairs gets larger, the StdDiff(random,Npairs) becomes smaller.We use Z is to score clusters in a dataset by statistical significance. A cluster with a smaller absolute meanDiff can have a higher absolute Z than a cluster with a larger absolute meanDiff if there are more pairs in the first cluster.
- By convention, we want the first molecule in the pair to have the higher activity, so we may reassign molecules in all pairs in the cluster as A and B so that meanDiff and Z is positive.
- Clusters can be written in order of decreasing Z, decreasing meanDiff, or decreasing Npairs. The default of T-ANALYZE is to sort by decreasing Z.
3.6. Selection of Statistically Significant Clusters
3.7. Selection of Clusters
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Allenmark, S. Chromatographic Enantioseparation. Methods and Applications; Ellis Horwood Publisher: Chichester, UK, 1988. [Google Scholar]
- Pirkle, W.H.; Pochapsky, T.C. Considerations of chiral recognition relevant to the liquid chromatography separation of enantiomers. Chem. Rev. 1989, 89, 347–362. [Google Scholar] [CrossRef]
- Okamoto, Y.; Yashima, E. Polysaccharide derivatives for chromatographic separation of enantiomers. Angew. Chem. Int. Ed. 1998, 37, 1021–1043. [Google Scholar] [CrossRef]
- Armstrong, D.W.; Han, S.M. Enantiomeric separations in chromatography. Crit. Rev. Anal. Chem. 1988, 19, 175–224. [Google Scholar] [CrossRef]
- Francotte, E.R. Enantioselective chromatography. An essential and versatile tool for the analytical and preparative separation of enantiomers. Chimia 1997, 51, 717–725. [Google Scholar]
- Allenmark, S.; Schurig, V. Chromatography on chiral stationary phases. J. Mat. Chem. 1997, 7, 1955–1963. [Google Scholar] [CrossRef]
- Lammerhofer, M.; Lindner, W. Liquid chromatographic enantiomer separation and chiral recognition by cinchona alkaloid-derived enantioselective separation materials. Adv. Chromatogr. 2008, 46, 1–107. [Google Scholar] [PubMed]
- Suzuki, T.; Timofei, S.; Iuoras, B.E.; Uray, G.; Verdino, P.; Fabian, W.M.F. Quantitative structure–enantioselective retention relationships for chromatographic separation of arylalkylcarbinols on Pirkle type chiral stationary phases. J. Chromatogr. A 2001, 922, 13–23. [Google Scholar] [CrossRef]
- Carotti, A.; Altomare, C.; Cellamare, S.; Monforte, A.M.; Bettoni, G.; Loiodice, F.; Tangari, N.; Tortorella, V. LFER and CoMFA studies on optical resolution of -alkyl –aryloxyacetic acid methyl esters on DACH-DNB chiral stationary phase. J. Comput. Aided Mol. Des. 1995, 9, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Fabian, W.M.F.; Stamfer, W.; Mazur, M.; Uray, G. Modeling the Chromatographic Enantioseparation of Aryl- and Hetarylcarbinols on ULMO, a Brush-Type Chiral Stationary Phase, by 3D-QSAR Techniques. Chirality 2003, 15, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Del Rio, A.; Piras, P.; Roussel, C. Enantiophore Modeling in 3D-QSAR. A Data Mining Application on Whelk-O1 Chiral Stationary Phase. Chirality 2006, 18, 498–508. [Google Scholar] [PubMed]
- Sheridan, R.; Schafer, W.; Piras, P.; Zawatzky, K.; Sherer, E.C.; Roussel, C.; Welch, C.J. Toward structure-based predictive tools for the selection of chiral stationary phases for the chromatographic separation of enantiomers. J. Chromatogr. A. 2016. [Google Scholar] [CrossRef] [PubMed]
- Roussel, C.; Piras, P. Chirbase: A molecular database for storage and retrieval of chromatographic chiral separations. Pure Appl. Chem. 1993, 65, 235–244. [Google Scholar] [CrossRef]
- Dossetter, A.G.; Griffen, E.J.; Leach, A.G. Matched molecular pair analysis in drug discovery. Drug Discov. Today 2013, 18, 724–731. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.H.; Liu, X.L.; Wu, Z.J.; Du, X.L.; Zhang, X.M.; Yuan, W.C. Organocatalytic Asymmetric Conjugate Addition of 3-Monosubstituted Oxindoles to (E)-1,4-Diaryl-2-buten-1,4-diones: A Strategy for the Indirect Enantioselective Furanylation and Pyrrolylation of 3-Alkyloxindoles. Chem. Eur. J. 2012, 18, 6679–6687. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, T.; Omote, O.; Maruoka, K. Asymmetric Alkynylation of C1-Substituted C,N-Cyclic Azomethine Imines by CuI/Chiral Brønsted Acid Co-Catalyst. Angew. Chem. Int. Ed. 2011, 50, 8952–8955. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Xu, F.; Lin, X.; Wang, Y. Highly Enantioselective Pictet–Spengler Reaction Catalyzed by SPINOL-Phosphoric Acids. Chem. Eur. J. 2012, 18, 3148–3152. [Google Scholar] [CrossRef] [PubMed]
- Ye, K.Y.; Lynam, K.G.; Stringham, R.W. Effect of amine mobile phase additives on chiral subcritical fluid chromatography using polysaccharide stationary phases. J. Chromatogr. A 2004, 1041, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Ye, K.Y.; Lord, B.; Stringham, R.W. Memory effect of mobile phase additives in chiral separations on a Chiralpak AD column. J. Chromatogr. A 2002, 945, 139–146. [Google Scholar] [CrossRef]
- Wu, Z.; Perez, M.; Scalone, M.; Ayad, T.; Ratovelomanana-Vidal, V. Ruthenium-Catalyzed Asymmetric Transfer Hydrogenation of 1-Aryl-Substituted Dihydroisoquinolines: Access to Valuable Chiral 1-Aryl-Tetrahydroisoquinoline Scaffolds. Angew. Chem. Int. Ed. 2013, 52, 4925–4928. [Google Scholar] [CrossRef] [PubMed]
- Ye, K.Y.; Stringham, R. Effect of mobile phase amine additives on enantioselectivity for phenylalanine analogs. J. Chromatogr. A 2001, 927, 53–60. [Google Scholar] [CrossRef]
- Andrus, M.B.; Christiansen, M.A.; Hicken, E.J.; Gainer, M.J.; Bedke, D.K.; Harper, K.C.; Mikkelson, S.R.; Dodson, D.S.; Harris, D.T. Phase-Transfer-Catalyzed Asymmetric Acylimidazole Alkylation. Org. Lett. 2007, 9, 4865–4868. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Yu, J.; Sun, X.X.; Rao, Q.Q.; Gong, L.Z. Organocatalytic Asymmetric Three-Component Cyclization of Cinnamaldehydes and Primary Amines with 1,3-Dicarbonyl Compounds: Straightforward Access to Enantiomerically Enriched Dihydropyridines. Angew. Chem. Int. Ed. 2008, 47, 2458–2462. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, Y.; Gridnev, I.D.; Ikariya, T. Enantioselective and Z/E-Selective Conjugate Addition of α-Substituted Cyanoacetates to Acetylenic Esters Catalyzed by Bifunctional Ruthenium and Iridium Complexes. Angew. Chem. Int. Ed. 2010, 49, 8157–8160. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, Y.; Watanabe, M.; Gridnev, I.D.; Ikariya, T. Enantioselective Direct Amination of α-Cyanoacetates Catalyzed by Bifunctional Chiral Ru and Ir Amido Complexes. J. Am. Chem. Soc. 2008, 130, 2158–2159. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Zhang, Y.; Staples, R.J.; Huang, R.; Wulff, W.D. Double Stereodifferentiation in the Catalytic Asymmetric Aziridination of Imines Prepared from α-Chiral Amines. Chem. Eur. J. 2012, 18, 5302–5313. [Google Scholar] [CrossRef] [PubMed]
- Navarre, L.; Martinez, R.; Genet, J.P.; Darses, S. Access to Enantioenriched α-Amino Esters via Rhodium-Catalyzed 1,4-Addition/Enantioselective Protonation. J. Am. Chem. Soc. 2008, 130, 6159–6169. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, J.A.; Stringham, R.W.; Xiang, D.; Waltemire, R.E. Empirical relationship between chiral selectivity and mobile phase modifier properties. J. Chromatogr. A 1999, 852, 383–394. [Google Scholar] [CrossRef]
- Lipka, E.; Vaccher, C.; Bonte, J.P. Enantioseparation of benzoxazolinone aminoalcohols and their aminoketone precursors, potential adrenergic ligands, by analytical and preparative liquid chromatography on amylose chiral stationary phases and characterization of the enantiomers. Chirality 2009, 21, 769–776. [Google Scholar] [CrossRef] [PubMed]
- Sarac, S.; Tozkoparan, B.; Ertan, M. Separation of enantiomers of some condensed dihydropyrimidine derivatives by HPLC using chiral stationary phases. FABAD J. Pharm. Sci. 2000, 25, 145–151. [Google Scholar]
- Yarim, M.; Sarac, S. Separation of the enantiomers of 4-aryl-7,7-dimethyl- and 1,7,7-trimethyl-1,2,3,4,5,6,7,8-octahydroquinazoline-2,5-diones by chiral HPLC. Chromatographia 2002, 56, 307–312. [Google Scholar] [CrossRef]
- Tozkoparan, B.; Yarim, M.; Sarac, S.; Ertan, M.; Kelicen, P.; Altinok, G.; Demirdamar, R. Studies on synthesis, chromatographic resolution, and antiinflammatory activities of some 2-thioxo-1,2,3,4-tetrahydropyrimidines and their condensed derivatives. Arch. Pharm. Pharm. Med. Chem. 2000, 333, 415–420. [Google Scholar] [CrossRef]
- Aboul-Enein, H.Y.; Abou-Basha, L.I.; Bakr, S.A. Direct enantioselective separation of some propranolol analogs by HPLC on normal and reversed cellulose chiral stationary phases. Chirality 1996, 8, 153–156. [Google Scholar] [CrossRef]
- Azzolina, O.; Vercesi, D.; Collina, S.; Ghislandi, V. Chiral resolution of methyl 2-aryloxypropionates by biocatalytic stereospecific hydrolysis. Farmaco 1995, 50, 221–226. [Google Scholar]
- Azzolina, O.; Collina, S.; Ghislandi, V. Optical resolution of aryloxypropionic acids and their esters by HPLC on cellulose tris-3, 5-dimethyl-triphenylcarbamate derivative. Farmaco 1993, 48, 1401–1416. [Google Scholar]
- Bargmann-Leyder, N.; Tambuté, A.; Caude, M. A comparison of LC and SFC for cellulose- and amylose-derived chiral stationary phases. Chirality 1995, 7, 311–325. [Google Scholar] [CrossRef]
- Cirilli, R.; Ferretti, R.; Gallinella, B.; Turchetto, L; Bolasco, A.; Secci, D.; Chimenti, P.; Pierini, M.; Fares, V.; Befani, O.; La Torre, F. Enantiomers of C5-chiral 1-acetyl-3,5-diphenyl-4,5-dihydro-(1H)-pyrazole derivatives: Analytical and semipreparative HPLC separation, chiroptical properties, absolute configuration, and inhibitory activity against monoamine oxidase. Chirality 2004, 16, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Miyazawa, T.; Shindo, Y.; Yamada, T. Separation of Enantiomers of N-Protected Non-Protein Amino Acid Esters by Chiral High-Performance Liquid Chromatography. Chromatographia 2004, 60, 419–424. [Google Scholar] [CrossRef]
- Okamoto, Y.; Kaida, Y.; Aburatani, R.; Hatada, K. Optical resolution of amino acid derivatives by high-performance liquid chromatography on tris(phenylcarbamate)s of cellulose and amylose. J. Chromatogr. 1989, 477, 367–376. [Google Scholar] [CrossRef]
- Miyazawa, T.; Hamahara, K.; Matsuoka, M.; Shindo, Y.; Yamada, T. Protease-catalyzed incorporation of non-protein amino acids into peptides via the kinetically controlled approach. Anal. Lett. 1993, 26, 457–473. [Google Scholar] [CrossRef]
- Wolf, R.M.; Francotte, E.; Lohmann, D. Quantitative correlation between calculated molecular properties and retention of a series of structurally related racemates on cellulose triacetate. J. Chem. Soc. Perkin Trans. 1988, 2, 893–901. [Google Scholar] [CrossRef]
- Wolf, R.M.; Francotte, E.; Hainmüller, J. Chiral discrimination of the enantiomers of δ-phenyl-δ-valerolactone by cellulose triacetate: A chromatographic and microcalorimetric study of the thermodynamics. Chirality 1993, 5, 538–544. [Google Scholar] [CrossRef]
- Francotte, and Ackermann Chirality and Biological Activity; Frank, H.; Holmstedt, B.; Testa, B. (Eds.) LISS A.R. INC: New York, NY, USA, 1990; pp. 63–70.
- Chilmonczyk, Z.; Ksycińska, H.; Lisowska-Kuźmicz, M.; Jończyk, A.; Mazgajska, M.; Jarończyk, M.; Strzelczyk, A.; Aboul-Enein, H.Y. Structure-enantioselectivity relationship of hypnotic-sedative 1,4-disubstituted piperazine derivatives on cellulose tris(4-methylbenzoate) chiral stationary phase. Anal. Chim. Acta 2005, 536, 7–13. [Google Scholar] [CrossRef]
- Chilmonczyk, Z.; Sienicki, Ł.; Łozowicka, B.; Lisowska-Kuźmicz, M.; Jończyk, A.; Aboul-Enein, H.Y. Structure-Retention Relationship in a Series of 1,4-Disubstituted Piperazine Derivatives on Carbohydrate Chiral Stationary Phases. Il Farmaco 2005, 60, 439–443. [Google Scholar] [CrossRef] [PubMed]
- Chilmonczyk, Z.; Ksycińska, H.; Mazgajska, M.; Cybulski, J.; Kaliszan, R. Non-linear structure-enantioselective retention relationships in a homologous series of 1,4-disubstituted piperazine derivatives. J. Chromatogr. A 1997, 788, 81–85. [Google Scholar] [CrossRef]
- Chen, C.A.; Wu, K.H.; Gau, H.M. Highly Enantioselective Aryl Additions of [AlAr3(thf)] to Ketones Catalyzed by a Titanium(IV) Catalyst of (S)-Binol. Angew. Chem., Int. Ed. 2007, 46, 5373–5376. [Google Scholar] [CrossRef] [PubMed]
- Shieh, W.C.; Cantrell, W.R.; Carlson, J.A. Asymmetric reduction of ortho-substituted benzophenones with B-chlorodiisopinocampheylborane: A convenient synthesis of enantiomerically enriched benzhydrols. Tetrahedron. Lett. 1995, 36, 3797–3800. [Google Scholar] [CrossRef]
- Pirkle, W.H.; Tsipouras, A. Direct liquid chromatographic separation of benzodiazepinone enantiomers. J. Chromatogr. 1984, 291, 291–298. [Google Scholar] [CrossRef]
- Azzolina, O.; Ghislandi, V.; Vercesi, D. Optical resolution of benzodiazepine esters by HPLC. Farmaco 1990, 45, 603–615. [Google Scholar] [PubMed]
- Lu, X.L.; Yang, S.K. Enantiomer resolution of camazepam and its derivatives and enantioselective metabolism of camazepam by human liver microsomes. J. Chromatogr. A 1994, 666, 249–257. [Google Scholar] [CrossRef]
- Lu, X.L.; Yang, S.K. N,N-dimethylcarbamyl derivatives of oxazepam. Chirality 1991, 3, 212–219. [Google Scholar]
- Pirkle, W.H.; Pochapsky, T.C.; Mahler, G.S.; Field, R.E. Chromatographic separation of the enantiomers of 2-carboalkoxyindolines and N-aryl-α-amino esters on chiral stationary phases derived from N-(3,5-dinitrobenzoyl)-α-amino acids. J. Chromatogr. 1985, 348, 89–96. [Google Scholar] [CrossRef]
- Lee, W.J. Chromatographic Separation of the Enantiomers of Amino Acid Esters as Benzophenone Imine Derivatives. Bull. Korean Chem. Soc. 1998, 19, 715–717. [Google Scholar]
- Job, G.E.; Shvets, A.; Pirkle, W.H.; Kuwahara, S.; Kosaka, M.; Kasai, Y.; Taji, H.; Fujita, K.; Watanabe, M.; Harada, N. The effects of aromatic substituents on the chromatographic enantioseparation of diarylmethyl esters with the Whelk-O1 chiral stationary phase. J. Chromatogr. A 2004, 1055, 41–53. [Google Scholar] [CrossRef]
- Hong, S.; Tian, S.; Metz, M.V.; Marks, T.J. C2-Symmetric Bis(oxazolinato)lanthanide Catalysts for Enantioselective Intramolecular Hydroamination/Cyclization. J. Am. Chem. Soc. 2003, 125, 14768–14783. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.S.; Marks, T.J.; McDonald, F.E. Organolanthanide-catalyzed intramolecular hydroamination/cyclization/bicyclization of sterically encumbered substrates. Scope, selectivity, and catalyst thermal stability for amine-tethered unactivated 1,2-disubstituted alkenes. J. Org. Chem. 2004, 69, 1038–1052. [Google Scholar] [CrossRef] [PubMed]
- Kleidernigg, O.P.; Kappe, C.O. Separation of enantiomers of 4-aryldihydropyrimidines by direct enantioselective HPLC. A critical comparison of chiral stationary phases. Tetrahedron Asymmetry 1997, 8, 2057–2067. [Google Scholar] [CrossRef]
- Chen, S. Effect of Steric Hindrance on the Resolution of the Enantiomers of Alkyl Isothiocyanate Derivatives of Amino Acids on a Teicoplanin CSP Using a Methanol-Based Mobile Phase. Chromatographia 2006, 63, 97–102. [Google Scholar] [CrossRef]
- Chen, S. The enantioseparation of amino acids on a teicoplanin chiral stationary phase using non-aqueous mobile phases after pre-column derivatization with sulfur-containing reagents: The considerations of mobile phase composition and analyte structure variation on resolution enhancement. Biomed. Chromatogr. 2006, 20, 718–728. [Google Scholar] [PubMed]
- Welch, C.J. The Evolution of Chiral Stationary Phase Design in the Pirkle Laboratories. J. Chromatogr. 1994, 666, 3–26. [Google Scholar] [CrossRef]
- Pirkle, W.H.; Welch, C.J. Effect of Superfluous Remote Polar Functionality on Chiral Recognition. J. Chromatogr. A 1992, 589, 45–51. [Google Scholar] [CrossRef]
- Pirkle, W.H.; Hyun, M.H.; Tsipouras, A.; Hamper, B.C.; Banks, B. A rational approach to the design of highly effective chiral stationary phases for the liquid chromatographic separation of enantiomers. J. Pharm. Biomed. Anal. 1984, 2, 173–181. [Google Scholar] [CrossRef]
- Pirkle, W.H.; Däppen, R. Reciprocity in Chiral Recognition: Comparison of several chiral stationary phases. J. Chromatogr. A 1987, 404, 107–115. [Google Scholar] [CrossRef]
- Leblanc, Y.; Dufresne, C.; Carson, R.; Morency, L.; Welch, C.J. Enhanced Chromatographic Resolution of Alcohol Enantiomers as Phophate or Phosphonate Derivatives. Tetrahedron Asymmetry 2001, 12, 3063–3066. [Google Scholar] [CrossRef]
- Dai, Z.; Pittman, C.U., Jr.; Li, T. Enantiomeric Recognition of Racemic 4-Aryl-1,4-dihydropyridine Derivatives via Chiralpak AD-H Stationary Phases. Chirality 2012, 24, 854–859. [Google Scholar] [CrossRef] [PubMed]
- Chiral recognition by enantioselective liquid chromatography: Mechanisms and modern chiral stationary phases, Michael Lämmerhofer. J. Chromatogr. A 2010, 1217, 814–856.
- Toshihiko, H. Quantitative in Silico Chromatography: Computational Modelling of Molecular Interactions; Royal Society of Chemistry: London, UK, 2014. [Google Scholar]
- Gong, X.; Craven, T.; Schafer, W.; Pirzada, Z.; Biba, M.; Welch, C.J. Estimating Chromatographic Enantioselectivity (α) from Gradient Chiral Chromatography Data. Chirality 2011, 23, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Pirkle, W.H.; Welch, C.J. A Convenient Void Volume Marker for Several Chiral HPLC Columns. J. Liq. Chromatogr. 1991, 14, 1–8. [Google Scholar] [CrossRef]
- Sheridan, R.P.; Hunt, P.; Culberson, J.C. Molecular transformation as a way of finding and exploiting consistent local QSAR. J. Chem. Inf. Model. 2006, 46, 180–192. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds are not available from the authors.

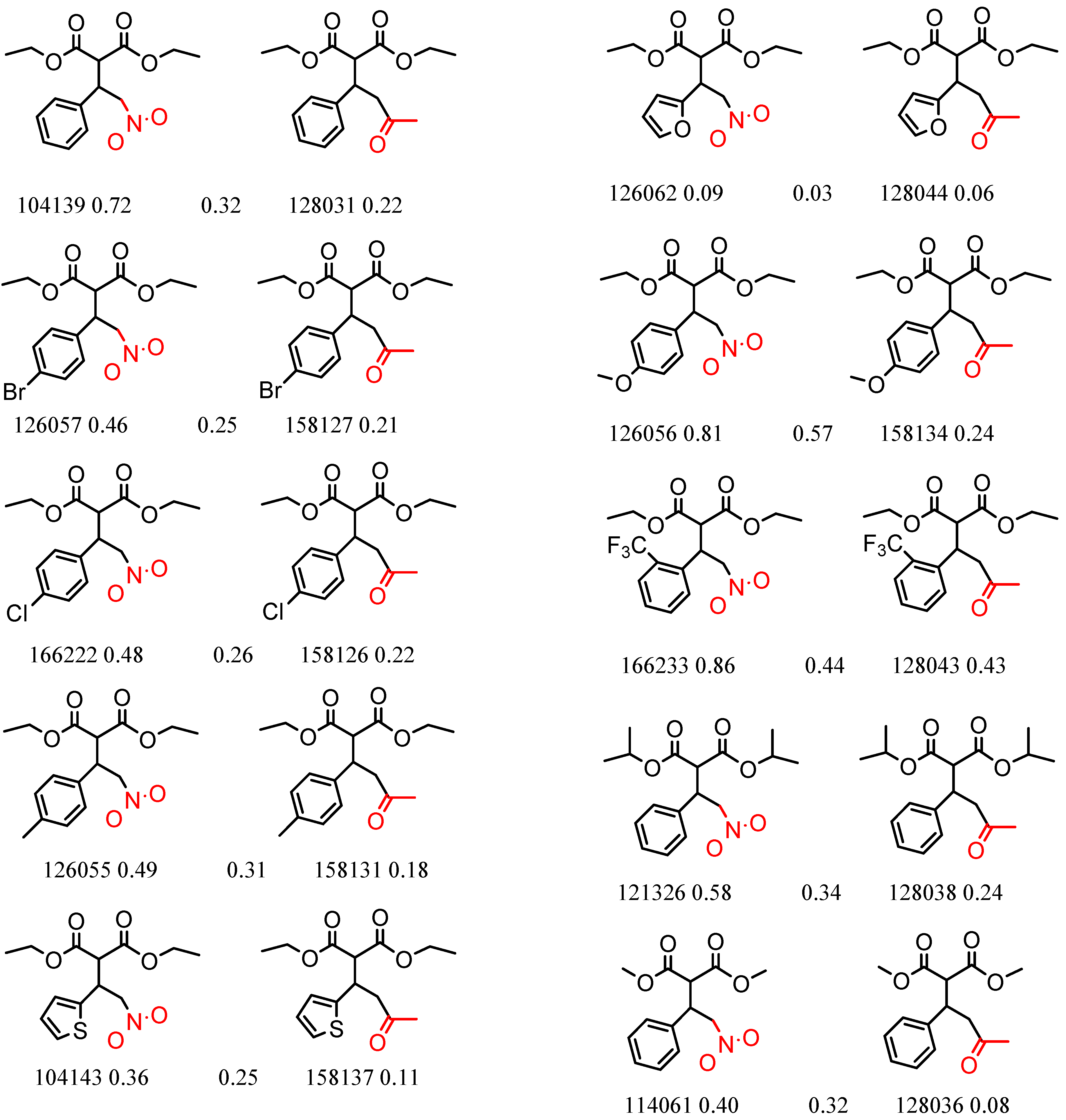
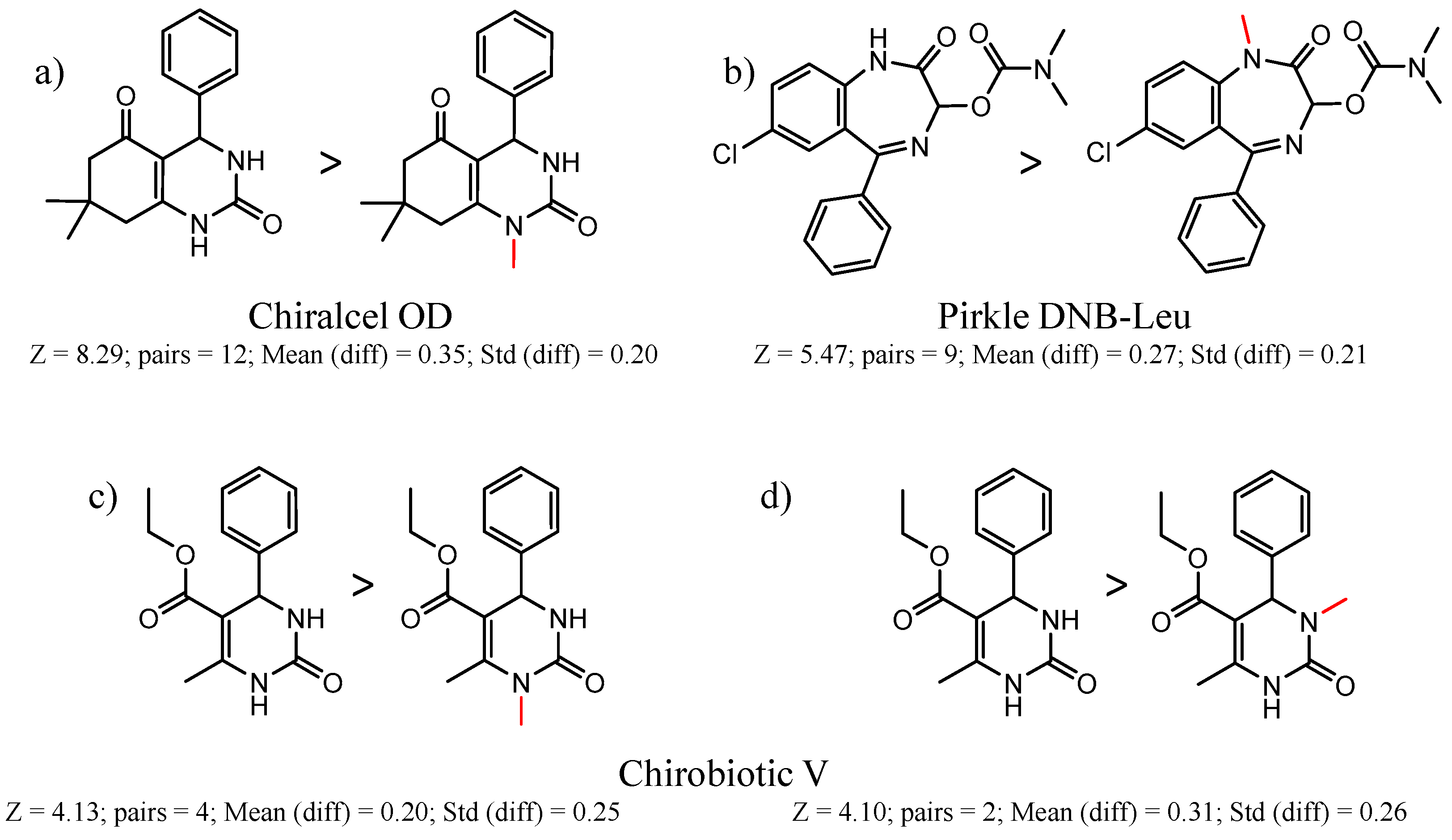
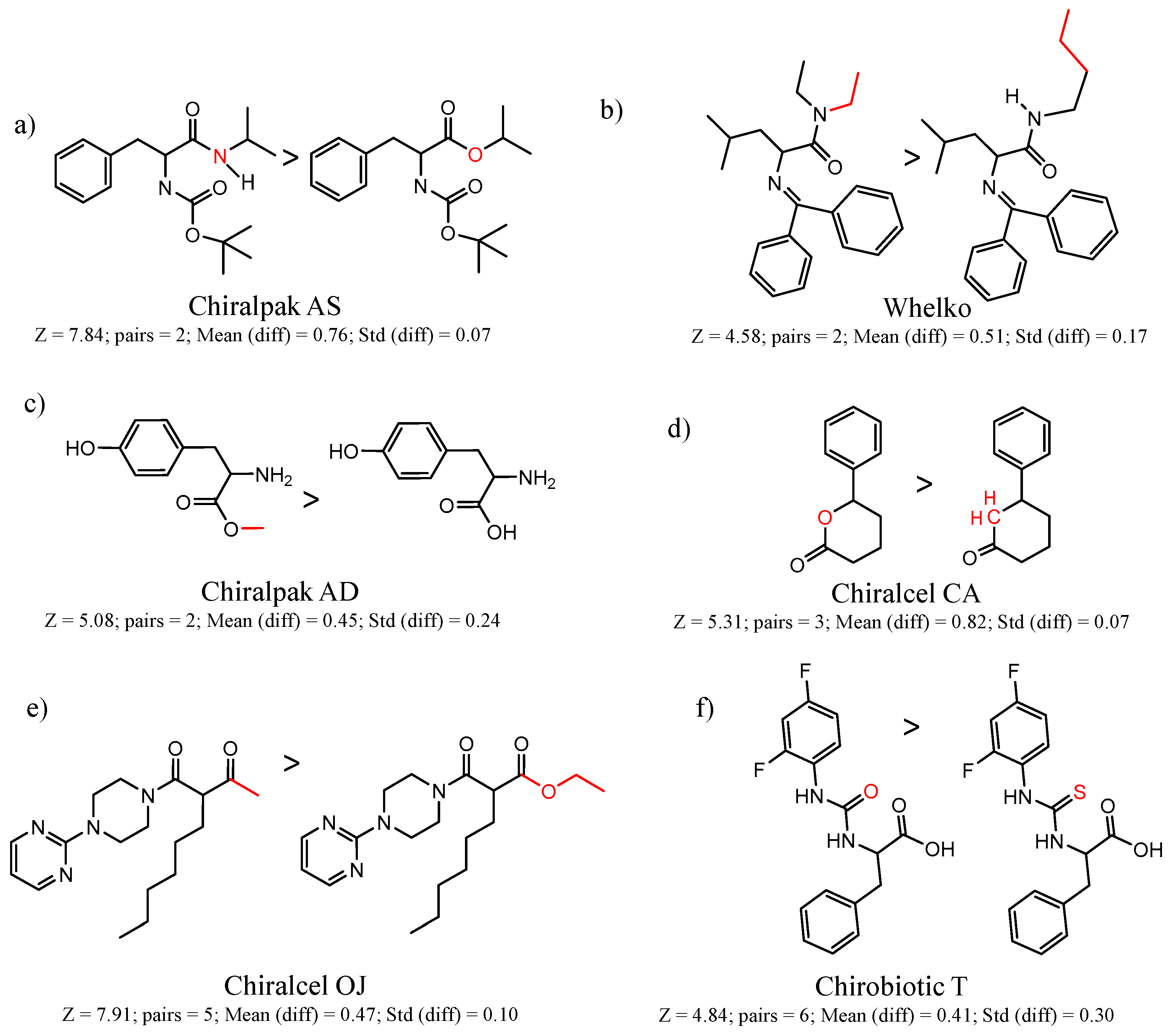
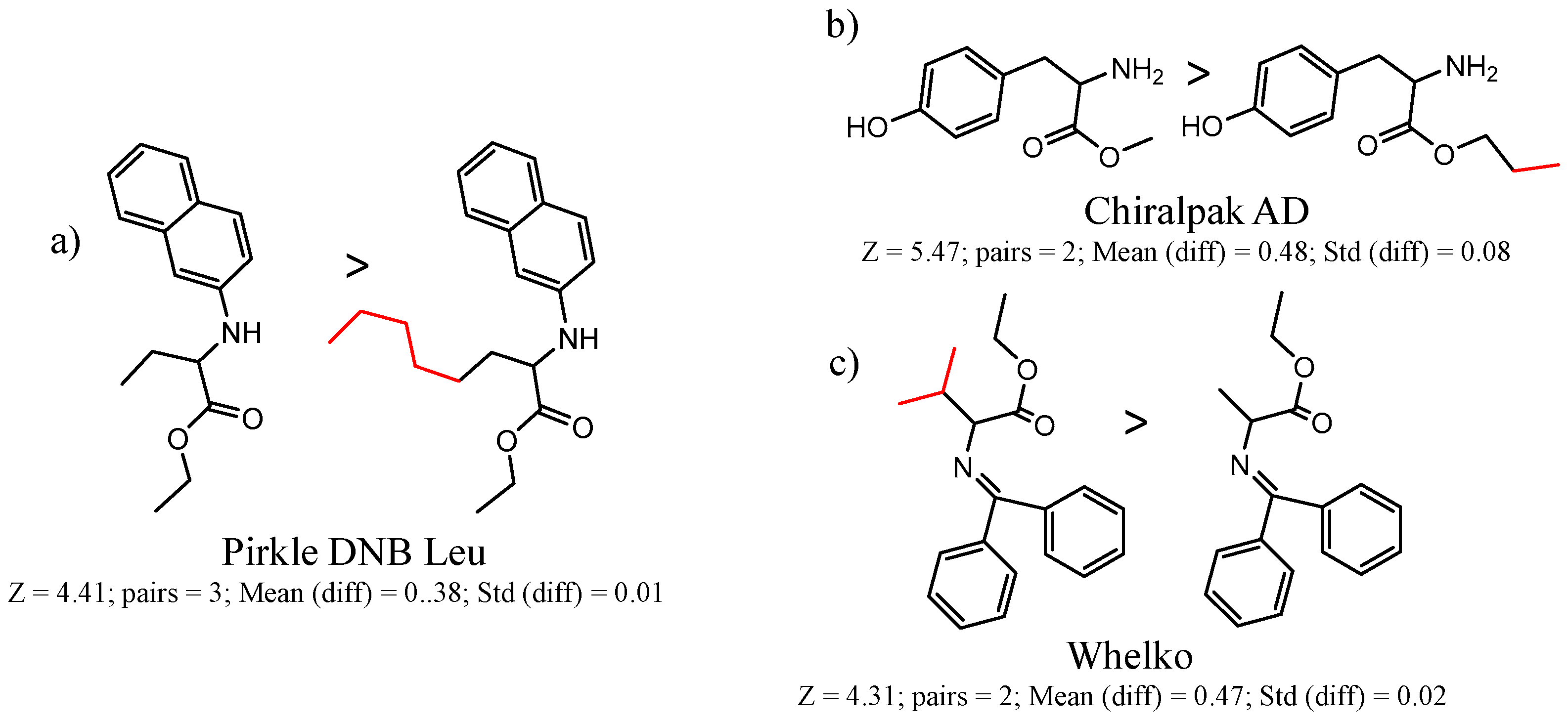
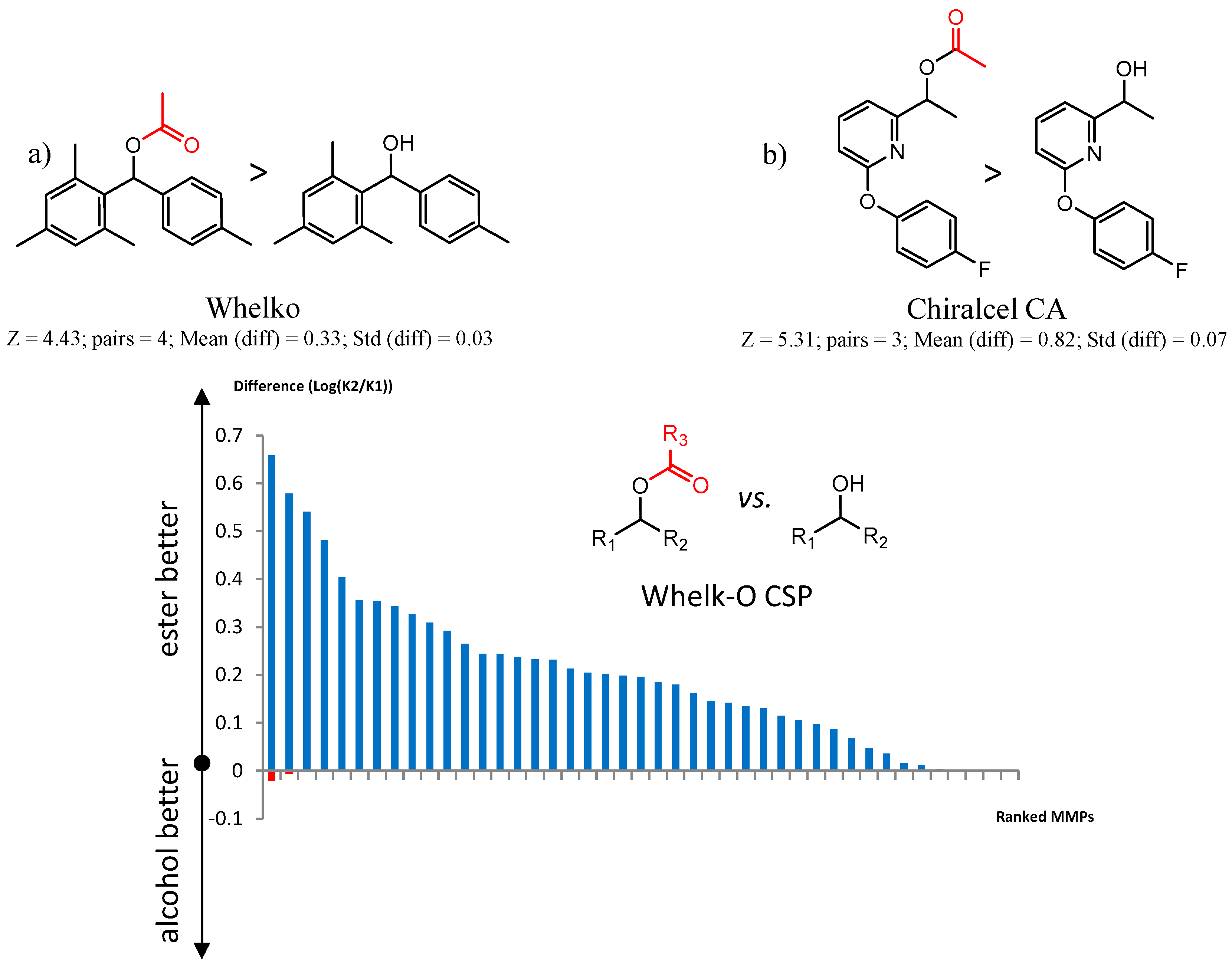



| CSP_NO | Description | Unique Molecules | Mean α | Stdev α | Total Clusters | Expected maxZ (95th Percentile) | Number of Significant Clusters |
|---|---|---|---|---|---|---|---|
| 394 | Chiral-AGP | 574 | 1.65 | 0.90 | 519 | 3.9 | 0 |
| 2966 | Crownpak-CR(+) 1 | 346 | 2.03 | 1.52 | 416 | 3.8 | 4 |
| 3575 | Ultron-ES-OVM | 190 | 1.55 | 0.59 | 90 | 3.4 | 0 |
| 15723 | Chirobiotic-R | 462 | 1.93 | 1.51 | 636 | 3.9 | 0 |
| 23735 | Chiralpak-AD | 11167 | 1.45 | 0.64 | 8348 | 4.6 | 65 |
| 45167 | Chiralpak-AS | 3665 | 1.48 | 0.68 | 4296 | 4.4 | 30 |
| 45172 | Chiralpak-IA | 1373 | 1.53 | 0.81 | 2099 | 4.2 | 11 |
| 45173 | Chiralcel-OD | 14350 | 1.47 | 0.64 | 22112 | 4.8 | 48 |
| 90201 | Chiralcel-CA-1 | 470 | 1.82 | 1.51 | 561 | 3.9 | 2 |
| 90220 | Chiralcel-OF | 291 | 1.40 | 0.63 | 257 | 3.7 | 4 |
| 90246 | Chiralcel-OJ | 4256 | 1.44 | 0.63 | 5108 | 4.4 | 20 |
| 90292 | (DNB)-Leu 1 | 610 | 1.71 | 1.17 | 1296 | 4.1 | 5 |
| 90589 | Whelk-O 1 | 1754 | 1.59 | 1.01 | 2246 | 4.2 | 14 |
| 90613 | Chiralcel-OZ | 314 | 1.53 | 0.76 | 204 | 3.6 | 0 |
| 90704 | Chirobiotic-V | 351 | 1.37 | 0.73 | 411 | 3.8 | 16 |
| 90752 | Chiralpak-AY | 216 | 1.67 | 0.99 | 108 | 3.5 | 0 |
| 90879 | Chirobiotic-T 1 | 1154 | 2.20 | 1.96 | 1920 | 4.2 | 20 |
| 91119 | Chirobiotic-TAG | 308 | 1.87 | 1.59 | 329 | 3.8 | 7 |
| 91423 | Chiralpak-IB | 680 | 1.39 | 0.62 | 876 | 4.0 | 11 |
| CSP_NO | Cluster | Z | No. Pairs | Mean (Diff) | Std (Diff) | Example | Reference |
|---|---|---|---|---|---|---|---|
| 23735 Chiralpak-AD | 1 | 11.52 | 12 | 0.43 | 0.23 |  | [15] |
| 5 | 6.93 | 6 | 0.35 | 0.03 |  | [16] | |
| 27 | 5.62 | 2 | 0.49 | 0.24 |  | [17] | |
| 28 | 5.61 | 2 | 0.49 | 0.27 |  | [17] | |
| 35 | 5.47 | 2 | 0.48 | 0.08 |  | [18,19] | |
| 44 | 5.14 | 2 | 0.46 | 0.07 |  | [18,19] | |
| 45 | 5.10 | 5 | 0.29 | 0.24 |  | [20] | |
| 46 | 5.08 | 2 | 0.45 | 0.24 |  | [19,21] | |
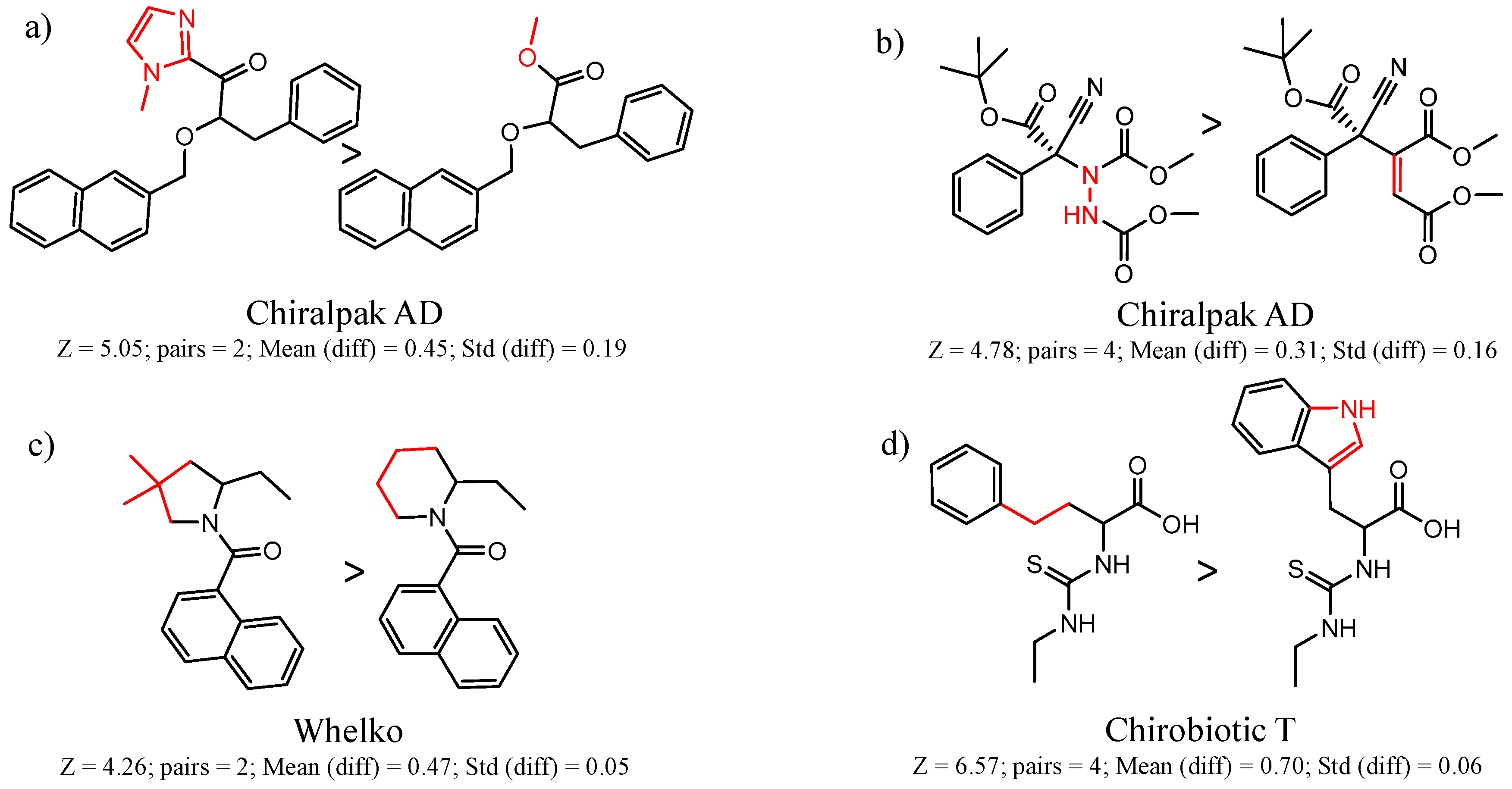
| 47 | 5.05 | 2 | 0.45 | 0.19 |  | [22] | |
| 61 | 4.80 | 4 | 0.31 | 0.01 |  | [23] | |
| 65 | 4.78 | 4 | 0.31 | 0.16 |  | [24,25] | |
| 45167 Chiralpak-AS | 1 | 7.84 | 2 | 0.76 | 0.07 |  | [26,27,28] |
| 28 | 4.57 | 2 | 0.44 | 0.29 |  | [29] | |
| 45173 Chiralcel-OD | 1 | 8.36 | 2 | 0.90 | 0.01 |  | [30] |
| 2 | 8.29 | 12 | 0.35 | 0.20 |  | [31] | |
| 3 | 8.05 | 2 | 0.86 | 0.08 |  | [30] | |
| 5 | 7.27 | 4 | 0.54 | 0.08 |  | [30] | |
| 6 | 7.00 | 4 | 0.52 | 0.09 |  | [30] | |
| 7 | 6.86 | 4 | 0.51 | 0.05 |  | [30] | |
| 9 | 6.71 | 3 | 0.55 | 0.40 |  | [30,32] | |
| 11 | 6.65 | 2 | 0.71 | 0.00 |  | [33] | |
| 15 | 6.17 | 3 | 0.51 | 0.09 |  | [34,35] | |
| 16 | 5.86 | 2 | 0.62 | 0.06 |  | [36,37] | |
| 17 | 5.85 | 2 | 0.62 | 0.06 |  | [30] | |
| 18 | 5.82 | 2 | 0.62 | 0.06 |  | [30] | |
| 19 | 5.76 | 3 | 0.48 | 0.13 |  | [30,34,35] | |
| 20 | 5.72 | 2 | 0.61 | 0.29 |  | [37] | |
| 23 | 5.61 | 2 | 0.60 | 0.26 |  | [37] | |
| 25 | 5.49 | 2 | 0.58 | 0.46 |  | [30,32] | |
| 27 | 5.43 | 2 | 0.57 | 0.01 |  | [30,32] | |
| 28 | 5.43 | 2 | 0.58 | 0.06 |  | [30] | |
| 33 | 5.11 | 2 | 0.55 | 0.01 |  | [30] | |
| 35 | 5.10 | 2 | 0.54 | 0.03 |  | [30,32] | |
| 37 | 5.07 | 2 | 0.54 | 0.31 |  | [37] | |
| 44 | 4.95 | 3 | 0.41 | 0.08 |  | [38,39,40] | |
| 47 | 4.82 | 2 | 0.52 | 0.01 |  | [30] | |
| 90201 Chiralcel-CA-1 | 1 | 5.31 | 3 | 0.82 | 0.07 |  | [41,42] |
| 2 | 4.25 | 8 | 0.42 | 0.29 |  | [43] | |
| 90246 Chiralcel-OJ | 1 | 7.91 | 5 | 0.47 | 0.10 |  | [44,45,46] |
| 16 | 4.59 | 2 | 0.45 | 0.00 |  | [47,48] | |
| 19 | 4.45 | 2 | 0.43 | 0.07 |  | [37] | |
| 90292 Pirkle DNB-Leu | 1 | 5.47 | 9 | 0.27 | 0.21 |  | [49,50,51,52] |
| 4 | 4.41 | 3 | 0.38 | 0.01 |  | [53] | |
| 90589 Whelk-O | 10 | 4.58 | 2 | 0.51 | 0.17 |  | [54] |
| 12 | 4.43 | 4 | 0.33 | 0.03 |  | [55] | |
| 13 | 4.31 | 2 | 0.47 | 0.02 |  | [54] | |
| 14 | 4.26 | 2 | 0.47 | 0.05 |  | [56,57] | |
| 90704 Chirobiotic-V | 12 | 4.52 | 2 | 0.34 | 0.34 |  | [58] |
| 13 | 4.30 | 2 | 0.33 | 0.36 |  | [58] | |
| 15 | 4.13 | 4 | 0.20 | 0.25 |  | [58] | |
| 16 | 4.10 | 2 | 0.31 | 0.26 |  | [58] | |
| 90879 Chirobiotic-T | 1 | 6.57 | 4 | 0.70 | 0.06 |  | [59] |
| 90879 | 3 | 5.29 | 2 | 0.85 | 0.02 |  | [59] |
| 90879 | 4 | 4.84 | 6 | 0.41 | 0.30 |  | [60] |
| 90879 | 5 | 4.76 | 2 | 0.77 | 0.17 |  | [60] |
| 90879 | 6 | 4.73 | 2 | 0.76 | 0.05 |  | [59] |
| 90879 | 10 | 4.62 | 4 | 0.49 | 0.33 |  | [58] |
| 90879 | 15 | 4.48 | 2 | 0.72 | 0.01 |  | [59] |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sheridan, R.P.; Piras, P.; Sherer, E.C.; Roussel, C.; Pirkle, W.H.; Welch, C.J. Mining Chromatographic Enantioseparation Data Using Matched Molecular Pair Analysis. Molecules 2016, 21, 1297. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules21101297
Sheridan RP, Piras P, Sherer EC, Roussel C, Pirkle WH, Welch CJ. Mining Chromatographic Enantioseparation Data Using Matched Molecular Pair Analysis. Molecules. 2016; 21(10):1297. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules21101297
Chicago/Turabian StyleSheridan, Robert P., Patrick Piras, Edward C. Sherer, Christian Roussel, William H. Pirkle, and Christopher J. Welch. 2016. "Mining Chromatographic Enantioseparation Data Using Matched Molecular Pair Analysis" Molecules 21, no. 10: 1297. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules21101297





