Isolation of α-Amylase Inhibitors from Kadsura longipedunculata Using a High-Speed Counter-Current Chromatography Target Guided by Centrifugal Ultrafiltration with LC-MS
Abstract
:1. Introduction
2. Results and Discussion
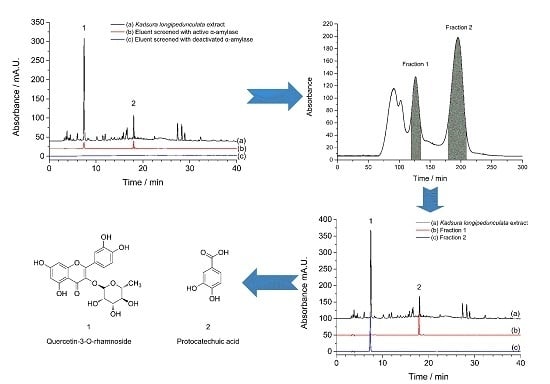
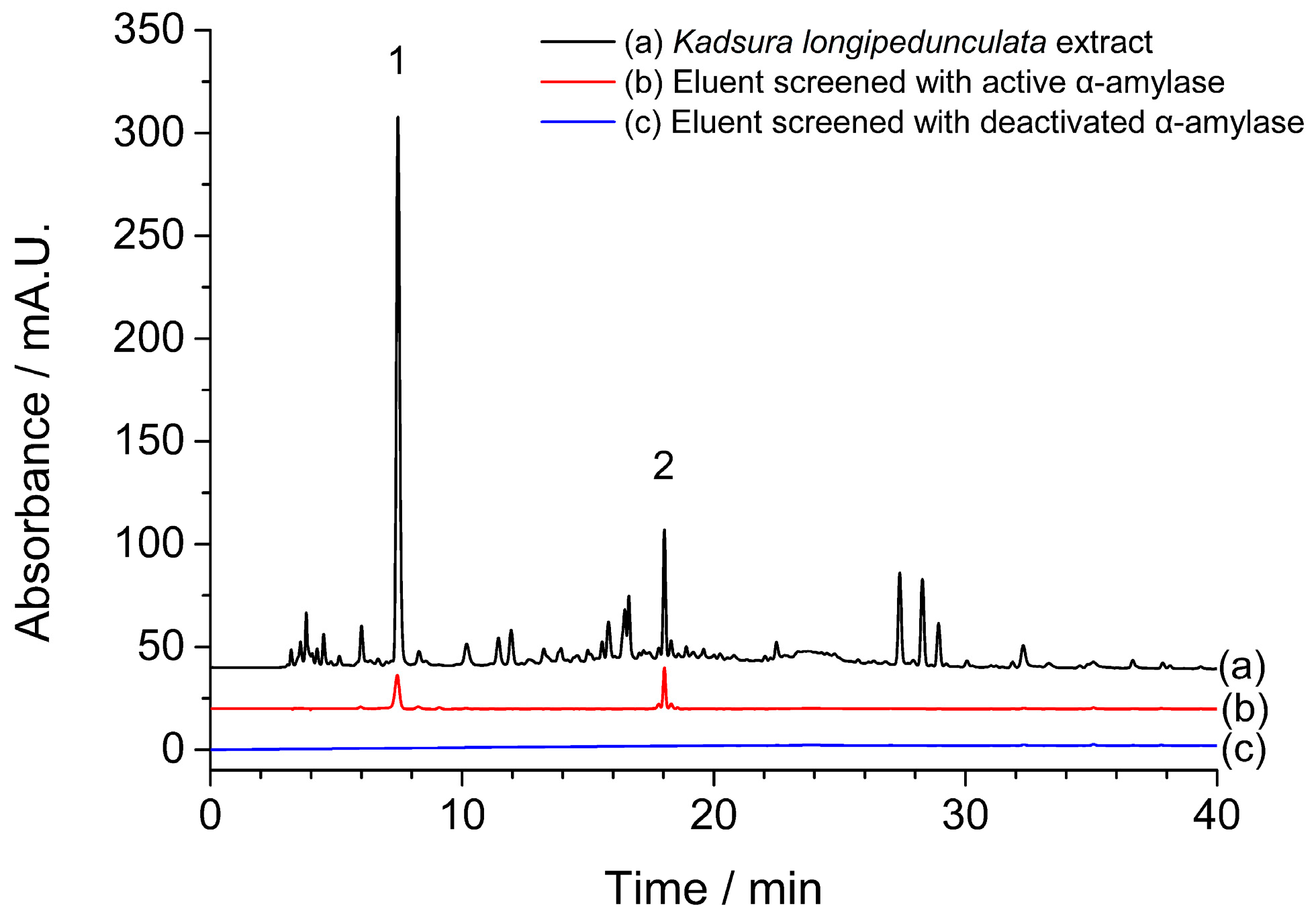
2.1. Screening of α-Amylase Inhibitors from Kadsura longipedunculata
2.2. Optimization of the HSCCC Solvent System
2.3. HSCCC Target Guided Separation
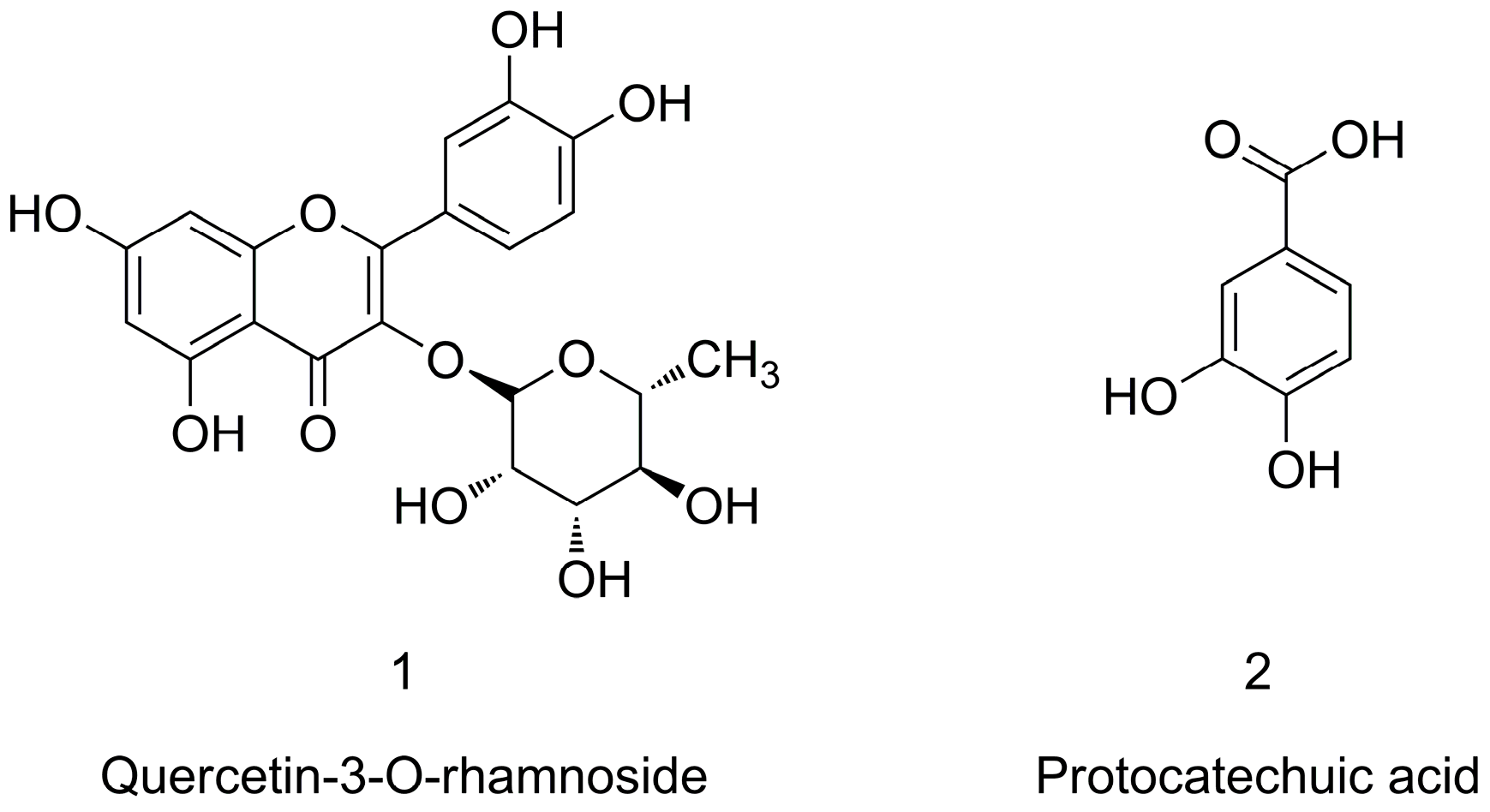
2.4. Identification of Target Compounds
2.5. Inhibition Activity on α-Amylase of Target Compounds
3. Materials and Methods
3.1. Materials
3.2. Preparation of Kadsura longipedunculata Extracts
3.3. HPLC Analysis Conditions
3.4. Screening of α-Amylase Inhibitors from Kadsura longipedunculata
3.5. HSCCC Target Guided Separation
3.6. Identification of α-Amylase Inhibitors
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural Products As Sources of New Drugs over the 30 Years from 1981 to 2010. J. Nat. Prod. 2012, 75, 311–335. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Ma, Q.; Ye, L.; Piao, G. The Traditional Medicine and Modern Medicine from Natural Products. Molecules 2016, 21, 559. [Google Scholar] [CrossRef] [PubMed]
- Nogawa, T.; Kamano, Y.; Yamashita, A.; Pettit, G.R. Isolation and Structure of Five New Cancer Cell Growth Inhibitory Bufadienolides from the Chinese Traditional Drug Ch’an Su. J. Nat. Prod. 2001, 64, 1148–1152. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, X.; Gao, X.; Nie, X.; Yang, Y.; Fan, X. Development of fluorescence imaging-based assay for screening cardioprotective compounds from medicinal plants. Anal. Chim. Acta 2011, 702, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Luque-Garcia, J.L.; Neubert, T.A. Sample preparation for serum/plasma profiling and biomarker identification by mass spectrometry. J. Chromatogr. A 2007, 1153, 259–276. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Yu, R.; Ai, N.; Fan, X. Rapid screening natural-origin lipase inhibitors from hypolipidemic decoctions by ultrafiltration combined with liquid chromatography–mass spectrometry. J. Pharm. Biomed. 2015, 104, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Hwang, S.H.; Huang, B.; Lim, S.S. Identification of tyrosinase specific inhibitors from Xanthium strumarium fruit extract using ultrafiltration-high performance liquid chromatography. J. Chromatogr. B 2015, 1002, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Peng, M.; Liu, L.; Shi, S.; Peng, S. Screening, Identification, and Potential Interaction of Active Compounds from Eucommia ulmodies Leaves Binding with Bovine Serum Albumin. J. Agric. Food Chem. 2012, 60, 3119–3125. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Jermihov, K.; Nam, S.J.; Sturdy, M.; Maloney, K.; Qiu, X.; Chadwick, L.R.; Main, M.; Chen, S.N.; Mesecar, A.D.; et al. Screening Natural Products for Inhibitors of Quinone Reductase-2 Using Ultrafiltration LC-MS. Anal. Chem. 2011, 83, 1048–1052. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Jung, Y.; Kim, S.N. Identification of Eupatilin from Artemisia argyi as a Selective PPARα Agonist Using Affinity Selection Ultrafiltration LC-MS. Molecules 2015, 20, 13753–13763. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.L.; Qian, Z.M.; Luo, Y.D.; Tang, D.; Chen, H.; Yi, L.; Li, P. Screening and mechanism study of components targeting DNA from the Chinese herb Lonicera japonica by liquid chromatography/mass spectrometry and fluorescence spectroscopy. Biomed. Chromatogr. 2008, 22, 1164–1172. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xia, Y.; Lu, Y.; Liang, J. Screening of permeable compounds in Flos Lonicerae Japonicae with liposome using ultrafiltration and HPLC. J. Pharm. Biomed. 2011, 54, 406–410. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhang, X.; Ma, Z.; Zhang, M.; Sun, F. Characterization of Aromatase Binding Agents from the Dichloromethane Extract of Corydalis yanhusuo Using Ultrafiltration and Liquid Chromatography Tandem Mass Spectrometry. Molecules 2010, 15, 3556–3566. [Google Scholar] [CrossRef] [PubMed]
- Friesen, J.B.; McAlpine, J.B.; Chen, S.N.; Pauli, G.F. Countercurrent Separation of Natural Products: An Update. J. Nat. Prod. 2015, 78, 1765–1796. [Google Scholar] [CrossRef] [PubMed]
- Marston, A.; Hostettmann, K. Developments in the application of counter-current chromatography to plant analysis. J. Chromatogr. A 2006, 1112, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, I.A.; Fisher, D. Role of counter-current chromatography in the modernisation of Chinese herbal medicines. J. Chromatogr. A 2009, 1216, 740–753. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Li, Y.; Kipletting Tanui, E.; Han, L.; Jia, Y.; Zhang, L.; Wang, Y.; Zhang, X.; Zhang, Y. Fishing and knockout of bioactive compounds using a combination of high-speed counter-current chromatography (HSCCC) and preparative HPLC for evaluating the holistic efficacy and interaction of the components of Herba Epimedii. J. Ethnopharmacol. 2013, 147, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Zhang, Y.; Chen, W.; Cai, P.; Zhang, S.; Chen, X.; Shi, S. High-speed counter-current chromatography coupled online to high performance liquid chromatography–diode array detector–mass spectrometry for purification, analysis and identification of target compounds from natural products. J. Chromatogr. A 2015, 1385, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, Y.; Liu, Q.; Sun, C.; Li, J.; Yang, P.; Wang, X. Preparative Separation of Phenolic Compounds from Chimonanthus praecox Flowers by High-Speed Counter-Current Chromatography Using a Stepwise Elution Mode. Molecules 2016, 21, 1016. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, S.B.; Singhal, R.S. Conjugation of α-amylase with dextran for enhanced stability: Process details, kinetics and structural analysis. Carbohydr. Polym. 2012, 90, 1811–1817. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Husain, Q.; Ansari, S. Polyaniline-assisted silver nanoparticles: A novel support for the immobilization of α-amylase. Appl. Microbiol. Biot. 2013, 97, 1513–1522. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Cui, J.H.; Yin, T.; Sun, L.; Li, G. Activated effect of lignin on α-amylase. Food Chem. 2013, 141, 2229–2237. [Google Scholar] [CrossRef] [PubMed]
- Lordan, S.; Smyth, T.J.; Soler-Vila, A.; Stanton, C.; Ross, R.P. The α-amylase and α-glucosidase inhibitory effects of Irish seaweed extracts. Food Chem. 2013, 141, 2170–2176. [Google Scholar] [CrossRef] [PubMed]
- Phan, M.A.T.; Wang, J.; Tang, J.; Lee, Y.Z.; Ng, K. Evaluation of α-glucosidase inhibition potential of some flavonoids from Epimedium brevicornum. LWT-Food Sci. Technol. 2013, 53, 492–498. [Google Scholar] [CrossRef]
- Mulyaningsih, S.; Youns, M.; El-Readi, M.Z.; Ashour, M.L.; Nibret, E.; Sporer, F.; Herrmann, F.; Reichling, J.; Wink, M. Biological activity of the essential oil of Kadsura longipedunculata (Schisandraceae) and its major components. J. Pharm. Pharmacol. 2010, 62, 1037–1044. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wu, J.; Feng, X.; Jia, Y.; Huang, J.; Hao, Z.; Zhao, S.; Wang, J. In silico Analysis and Experimental Validation of Lignan Extracts from Kadsura longipedunculata for Potential 5-HT1AR Agonists. PLoS ONE 2015, 10, e0130055. [Google Scholar] [CrossRef] [PubMed]
- Zaugg, J.; Ebrahimi, S.N.; Smiesko, M.; Baburin, I.; Hering, S.; Hamburger, M. Identification of GABA A receptor modulators in Kadsura longipedunculata and assignment of absolute configurations by quantum-chemical ECD calculations. Phytochemistry 2011, 72, 2385–2395. [Google Scholar] [CrossRef] [PubMed]
- Gan, Z.; Liang, Z.; Chen, X.; Wen, X.; Wang, Y.; Li, M.; Ni, Y. Separation and preparation of 6-gingerol from molecular distillation residue of Yunnan ginger rhizomes by high-speed counter-current chromatography and the antioxidant activity of ginger oils in vitro. J. Chromatogr. B 2016, 1011, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, W.; Liu, Z. Preparative isolation, quantification and antioxidant activity of dihydrochalcones from Sweet Tea (Lithocarpus polystachyus Rehd.). J. Chromatogr. B 2015, 1002, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y. Golden rules and pitfalls in selecting optimum conditions for high-speed counter-current chromatography. J. Chromatogr. A 2005, 1065, 145–168. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Wang, L.; Gao, Y.; Zhao, B.; Wang, D.; Duan, W.; Yu, Z. Isolation of isochlorogenic acid isomers in flower buds of Lonicera japonica by high-speed counter-current chromatography and preparative high performance liquid chromatography. J. Chromatogr. B 2015, 981–982, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Gopi, K.; Anbarasu, K.; Renu, K.; Jayanthi, S.; Vishwanath, B.S.; Jayaraman, G. Quercetin-3-O-rhamnoside from Euphorbia hirta protects against snake Venom induced toxicity. BBA-Gen. Subj. 2016, 1860, 1528–1540. [Google Scholar] [CrossRef] [PubMed]
- Shobana, S.; Sreerama, Y.N.; Malleshi, N.G. Composition and enzyme inhibitory properties of finger millet (Eleusine coracana L.) seed coat phenolics: Mode of inhibition of α-glucosidase and pancreatic amylase. Food Chem. 2009, 115, 1268–1273. [Google Scholar] [CrossRef]
- Aderogba, M.; Ndhlala, A.; Rengasamy, K.; Van Staden, J. Antimicrobial and Selected in Vitro Enzyme Inhibitory Effects of Leaf Extracts, Flavonols and Indole Alkaloids Isolated from Croton menyharthii. Molecules 2013, 18, 12633–12644. [Google Scholar] [CrossRef] [PubMed]
- López-Martínez, L.M.; Santacruz-Ortega, H.; Navarro, R.-E.; Sotelo-Mundo, R.R.; González-Aguilar, G.A. A 1H-NMR Investigation of the Interaction between Phenolic Acids Found in Mango (Manguifera indica cv Ataulfo) and Papaya (Carica papaya cv Maradol) and 1,1-diphenyl-2-picrylhydrazyl (DPPH) Free Radicals. PLoS ONE 2015, 10, e0140242. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Hyun, T.K.; Kim, M.J. The inhibitory effects of ethanol extracts from sorghum, foxtail millet and proso millet on α-glucosidase and α-amylase activities. Food Chem. 2011, 124, 1647–1651. [Google Scholar] [CrossRef]
- Girish, T.K.; Pratape, V.M.; Prasada Rao, U.J.S. Nutrient distribution, phenolic acid composition, antioxidant and alpha-glucosidase inhibitory potentials of black gram (Vigna mungo L.) and its milled by-products. Food Res. Int. 2012, 46, 370–377. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the compounds quercetin-3-O-rhamnoside and protocatechuic acid are available from the authors.


| Solvent System | Ratio (v/v) | K Value | α | |
|---|---|---|---|---|
| Compound 1 | Compound 2 | |||
| n-Hexane–Ethyl acetate–Methanol–Water | 1:5:1:5 | 2.537 | 1.507 | 1.683 |
| 1.5:5:1.5:5 | 1.488 | 0.689 | 2.160 | |
| 2:5:2:5 | 0.912 | 0.358 | 2.547 | |
| 3:5:3:5 | 0.329 | 0.072 | 4.569 | |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cen, Y.; Xiao, A.; Chen, X.; Liu, L. Isolation of α-Amylase Inhibitors from Kadsura longipedunculata Using a High-Speed Counter-Current Chromatography Target Guided by Centrifugal Ultrafiltration with LC-MS. Molecules 2016, 21, 1190. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules21091190
Cen Y, Xiao A, Chen X, Liu L. Isolation of α-Amylase Inhibitors from Kadsura longipedunculata Using a High-Speed Counter-Current Chromatography Target Guided by Centrifugal Ultrafiltration with LC-MS. Molecules. 2016; 21(9):1190. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules21091190
Chicago/Turabian StyleCen, Yin, Aiping Xiao, Xiaoqing Chen, and Liangliang Liu. 2016. "Isolation of α-Amylase Inhibitors from Kadsura longipedunculata Using a High-Speed Counter-Current Chromatography Target Guided by Centrifugal Ultrafiltration with LC-MS" Molecules 21, no. 9: 1190. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules21091190