Characterization of Collagen Peptides in Elaphuri Davidiani Cornu Aqueous Extract with Proliferative Activity on Osteoblasts Using Nano-Liquid Chromatography in Tandem with Orbitrap Mass Spectrometry
Abstract
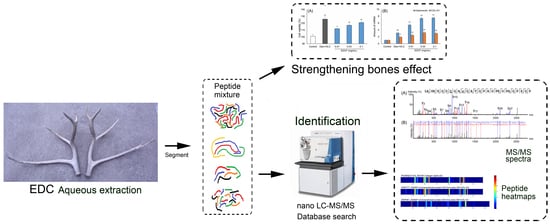
:1. Introduction
2. Results and Discussion
2.1. Inducing the Differentiation Activity of EDCF in Cultured Osteoblasts
2.2. Characterization of Peptides in EDCF
3. Materials and Methods
3.1. Chemicals and Materials
3.2. Preparation of EDC Extract and Ultrafiltration
3.3. Measurement of the Effects of EDC on Osteoblasts
3.3.1. Cell Culture
3.3.2. MTT Cell Viability Assay
3.3.3. Real Time Quantitative PCR
3.4. Peptide Characterization by Nano-LC-MS/MS
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jiang, Z.; Harris, R.B. Elaphurus Davidianus, The IUCN Red List of Threatened Species. Version 2016-3; Available online: http://www.iucnredlist.org (accessed on 21 December 2016).
- Li, F.T.; Duan, J.A.; Qian, D.W.; Guo, S.; Ding, Y.H.; Liu, X.H.; Qian, Y.F.; Peng, Y.R.; Ren, Y.J.; Chen, Y. Comparative analysis of nucleosides and nucleobases from different sections of Elaphuri Davidiani Cornu and Cervi Cornu by UHPLC–MS/MS. J. Pharm. Biomed. Anal. 2013, 83, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Meng, H.Y.; Qu, X.B.; Li, N.; Yuan, S.; Lin, Z. Effects of pilose antler and antler glue on osteoporosis of ovariectomized rats. J. Chin. Med. Mater. 2009, 32, 179–182. [Google Scholar]
- Qin, H.B.; Yang, C.Y.; Zhu, Q.; Zhu, Y.F. Effect of ethanol extract from Elaphuri Davidiani Cornu on anti-aging in mice. Chin. Tradit. Pat. Med. 2004, 26, 322–324. [Google Scholar]
- Yang, Z.Y.; Qin, H.B.; Cheng, H.L.; Zhu, Q. Effects of elk antlers ethanoic fluid extract on behavior and immune function of aging model mice. Chin. J. TCM Pharm. 2010, 25, 221–225. [Google Scholar]
- Guillerminet, F.; Beaupied, H.; Fabien-Soulé, V.; Tomé, D.; Benhamou, C.L.; Roux, C.; Blais, A. Hydrolyzed collagen improves bone metabolism and biomechanical parameters in ovariectomized mice: An in vitro and in vivo study. Bone 2010, 46, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Nomura, Y.; Oohashi, K.; Watanabe, M.; Kasugai, S. Increase in bone mineral density through oral administration of shark gelatin to ovariectomized rats. Nutrition 2005, 21, 1120–1126. [Google Scholar] [CrossRef] [PubMed]
- Rolf, H.J.; Enderle, A. Hard fallow deer antler: A living bone till antler casting? Anat. Rec. 1999, 255, 69–77. [Google Scholar] [CrossRef]
- Léonard, A.; Guiot, L.P.; Pirard, J.P.; Crine, M.; Balligand, M.; Blacher, S. Non-destructive characterization of deer (Cervus Elaphus) antlers by X-ray microtomography coupled with image analysis. J. Microsc. 2007, 225, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Buckley, M.; Collins, M.; Thomas-Oates, J.; Wilson, J.C. Species identification by analysis of bone collagen using matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2009, 23, 3843–3854. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Kim, M.G.; Leem, K.H. Osteogenic activity of collagen peptide via ERK/MAPK pathway mediated boosting of collagen synthesis and its therapeutic efficacy in osteoporotic bone by back-scattered electron imaging and microarchitecture analysis. Molecules 2013, 18, 15474–15489. [Google Scholar] [CrossRef] [PubMed]
- Wolters, D.A.; Washburn, M.P.; Yates, J.R., 3rd. An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 2001, 73, 5683–5690. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.H.; Yao, C.H.; Chan, T.M.; Huang, T.L.; Sen, Y.; Huang, C.Y.; Ho, C.Y. Effects of different concentrations of collagenous peptide from fish scales on osteoblast proliferation and osteoclast pesorption. Chin. J. Physiol. 2016, 59, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Bella, J. Collagen structure: New tricks from a very old dog. Biochem. J. 2016, 473, 1001–1025. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Wang, M.; Duan, J.A.; Guo, J.M.; Tang, Y.P. Purification and identification of three novel antioxidant peptides from Cornu Bubali (water buffalo horn). Peptides 2010, 31, 786–793. [Google Scholar] [CrossRef] [PubMed]
- Piovesana, S.; Capriotti, A.L.; Cavaliere, C.; La Barbera, G.; Samperi, R.; Zenezini Chiozzi, R.; Laganà, A. Peptidome characterization and bioactivity analysis of donkey milk. J. Proteom. 2015, 119, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Fujioka, M.; Sugimoto, K.; Mu, G.; Ishimi, Y. Assessment of effectiveness of oral administration of collagen peptide on bone metabolism in growing and mature rats. J. Bone Miner. Metab. 2004, 22, 547–553. [Google Scholar] [CrossRef] [PubMed]
- De Almeida Jackix, E.; Cúneo, F.; Amaya-Farfan, J.; de Assunção, J.V.; Quintaes, K.D. A food supplement of hydrolyzed collagen improves compositional and biodynamic characteristics of vertebrae in ovariectomized rats. J. Med. Food 2010, 13, 1385–1390. [Google Scholar] [CrossRef] [PubMed]
- Oesser, S.; Seifert, J. Stimulation of type II collagen biosynthesis and secretion in bovine chondrocytes cultured with degraded collagen. Cell Tissue Res. 2003, 311, 393–399. [Google Scholar] [PubMed]
- Iwai, K.; Hasegawa, T.; Taguchi, Y.; Morimatsu, F.; Sato, K.; Nakamura, Y.; Higashi, A.; Kido, Y.; Nakabo, Y.; Ohtsuki, K. Identification of food-derived collagen peptides in human blood after oral ingestion of gelatin hydrolysates. J. Agric. Food Chem. 2005, 53, 6531–6536. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.J.; Choi, R.C.; Cheung, A.W.; Chen, V.P.; Xu, S.L.; Dong, T.T.; Chen, J.J.; Tsim, K.W. Baicalin, a flavone, induces the differentiation of cultured osteoblasts. J. Biol. Chem. 2011, 286, 27882–27893. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, M.A.; Arnold, G.J.; Nynca, J.; Fröhlich, T.; Otte, K.; Ciereszko, A. Characterization of carp seminal plasma proteome in relation to blood plasma. J. Proteom. 2014, 98, 218–232. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of all compounds are available from the authors.





© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhai, Y.; Zhu, Z.; Zhu, Y.; Qian, D.; Liu, R.; Peng, Y.; Ding, Y.; Ouyang, Z.; Duan, J.-a. Characterization of Collagen Peptides in Elaphuri Davidiani Cornu Aqueous Extract with Proliferative Activity on Osteoblasts Using Nano-Liquid Chromatography in Tandem with Orbitrap Mass Spectrometry. Molecules 2017, 22, 166. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules22010166
Zhai Y, Zhu Z, Zhu Y, Qian D, Liu R, Peng Y, Ding Y, Ouyang Z, Duan J-a. Characterization of Collagen Peptides in Elaphuri Davidiani Cornu Aqueous Extract with Proliferative Activity on Osteoblasts Using Nano-Liquid Chromatography in Tandem with Orbitrap Mass Spectrometry. Molecules. 2017; 22(1):166. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules22010166
Chicago/Turabian StyleZhai, Yanjuan, Zhenhua Zhu, Yue Zhu, Dawei Qian, Rui Liu, Yunru Peng, Yuhua Ding, Zhen Ouyang, and Jin-ao Duan. 2017. "Characterization of Collagen Peptides in Elaphuri Davidiani Cornu Aqueous Extract with Proliferative Activity on Osteoblasts Using Nano-Liquid Chromatography in Tandem with Orbitrap Mass Spectrometry" Molecules 22, no. 1: 166. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules22010166