Antibacterial Synthetic Peptides Derived from Bovine Lactoferricin Exhibit Cytotoxic Effect against MDA-MB-468 and MDA-MB-231 Breast Cancer Cell Lines
Abstract
:1. Introduction
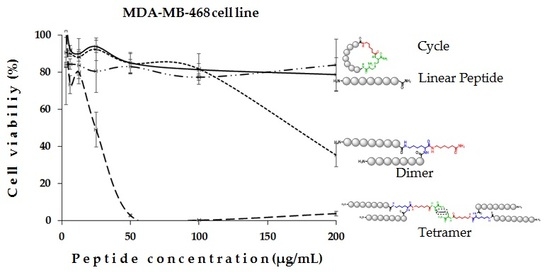
2. Results and Discussion
3. Materials and Methods
3.1. Reagents and Materials
3.2. Solid Phase Peptide Synthesis
3.3. LfcinB-Derived Peptide Characterization
3.3.1. Reverse Phase HPLC
3.3.2. Peptide Purification
3.3.3. MALDI-TOF MS
3.4. LfcinB-Derived Peptide Biological Activity
3.4.1. Antibacterial Activity Assays
3.4.2. MTT Assay
Acknowledgments
Author Contributions
Conflicts of Interest
References
- World Health Organization. Breast cancer. Available online: http://www.who.int/cancer/prevention/diagnosis-screening/breast-cancer/en/ (accessed on 23 July 2017).
- Richie, R.C.; Swanson, J.O. Breast Cancer: A Review of the Literature. J. Insur. Med. 2003, 35, 85–101. [Google Scholar] [PubMed]
- Satija, A.; Ahmed, S.M.; Gupta, R.; Ahmed, A.; Rana, S.P.; Singh, S.P.; Bhatnagar, S.M. Case report. Breast cancer pain management—A review of current & novel therapies. Indian J. Med. Res. 2014, 103, 216–225. [Google Scholar]
- Hormone Therapy for Breast Cancer. Available online: https://www.cancer.org/cancer/breast-cancer/treatment/hormone-therapy-for-breast-cancer.html (accessed on 23 July 2017).
- Dennison, S.; Whittaker, M.; Harris, F.; Phoenix, D. Anticancer alpha-helical peptides and structure/function relationships underpinning their interactions with tumour cell membranes. Curr. Protein Pept. Sci. 2006, 7, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Felício, M.R.; Silva, O.N.; Gonçalves, S.; Santos, N.C.; Franco, O.L. Peptides with Dual Antimicrobial and Anticancer Activities. Front. Chem. 2017, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Riedl, S.; Zweytick, D.; Lohner, K. Membrane-active host defense peptides—Challenges and perspectives for the development of novel anticancer drugs. Chem. Phys. Lipids 2011, 164, 766–781. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, J.A.; Kanwar, J.R.; Kanwar, R.K. Iron-free and iron-saturated bovine lactoferrin inhibit survivin expression and differentially modulate apoptosis in breast cancer. BMC Cancer 2015, 15, 425. [Google Scholar] [CrossRef] [PubMed]
- Farnaud, S.; Evans, R. Lactoferrin a multifunctional protein with antimicrobial properties. Mol. Immunol. 2003, 40, 395–405. [Google Scholar] [CrossRef]
- Lonnerdal, B.; Lyer, S. Lactoferrin: Molecular structure and biological function. Annu. Rev. Nutr. 1995, 15, 93–110. [Google Scholar] [CrossRef] [PubMed]
- García-Montoya, I.A.; Cendón, T.S.; Arévalo-Gallegos, S.; Rascón-Cruz, Q. Lactoferrin a multiple bioactive protein: An overview. Biochim. Biophys. Acta 2012, 1820, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Tomita, M.; Wakabayashi, H.; Shin, K.; Yamauchi, K.; Yaeshima, T.; Iwatsuki, K. Twenty-five years of research on bovine lactoferrin applications. Biochimie 2009, 91, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Bellamy, W.; Takase, M.; Yamauchi, K.; Wakabayashi, H.; Kawase, K.; Tomita, M. Identification of the bactericidal domain of lactoferrin. Biochim. Biophys. Acta 1992, 1121, 130–136. [Google Scholar] [CrossRef]
- Tomita, M.; Takase, M.; Wakabayashi, H.; Bellamy, W. Antimicrobial peptides of lactoferrin. In Lactoferrin: Structure Function; Hutchens, T.W., Rumball, S.V., Lönnerdal, B., Eds.; Plenum Press: New York, NY, USA, 1994; pp. 209–218. [Google Scholar]
- León, M.A.; Leal, A.L.; Almanzar, G.; Rosas, J.E.; García, J.E.; Rivera, Z.J. Antibacterial activity of synthetic peptides derived from Lactoferricin against Escherichia coli ATCC 25922 and Enterococcus faecalis ATCC 29212. Biomed. Res. Int. 2015, 2015, 1–8. [Google Scholar]
- Vorland, L.H.; Ulvatne, H.; Rekdal, O.; Svendsen, J.S. Initial binding sites of antimicrobial peptides in Staphylococcus aureus and Escherichia coli. Scand. J. Infect. Dis. 1999, 31, 467–473. [Google Scholar] [PubMed]
- Richardson, A.; de Antueno, R.; Duncan, R.; Hoskin, D.W. Intracellular delivery of bovine lactoferricin’s antimicrobial core (RRWQWR) kills T-leukemia cells. Biochem. Biophys. Res. Commun. 2009, 388, 736–741. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.R.; Chen, P.W.; Chen, Y.L.; Hsu, H.C.; Lin, C.C.; Chen, W.J. Bovine lactoferricin B induces apoptosis of human gastric cancer cell line AGS by inhibition of autophagy at a late stage. J. Dairy Sci. 2013, 96, 7511–7520. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Lönnerdal, B. Bovine lactoferrin and lactoferricin exert antitumor activities on human colorectal cancer cells (HT-29) by activating various signaling pathways. Biochem. Cell Biol. 2017, 95, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Iigo, M.; Kuhara, T.; Ushida, Y.; Sekine, K.; Moore, M.A.; Tsuda, H. Inhibitory effects of bovine lactoferrin on colon carcinoma 26 lung metastasis in mice. Clin. Exp. Metastasis 1999, 17, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Mader, J.S.; Salsman, J.; Conrad, D.M.; Hoskin, D.W. Bovine lactoferricin selectively induces apoptosis in human leukemia and carcinoma cell lines. Mol. Cancer Ther. 2005, 4, 612–624. [Google Scholar] [CrossRef] [PubMed]
- Yoo, Y.C.; Watanabe, S.; Watanabe, R.; Hata, K.; Shimazaki, K.; Azuma, I. Bovine lactoferrin and lactoferricin, a peptide derived from bovine lactoferrin, inhibit tumor metastasis in mice. Jpn. J. Cancer Res. 1997, 88, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Hilchie, A.L.; Vale, R.; Zemlak, T.S.; Hoskin, D.W. Generation of a hematologic malignancy-selective membranolytic peptide from the antimicrobial core (RRWQWR) of bovine lactoferricin. Exp. Mol. Pathol. 2013, 95, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Arias, M.; Hilchie, A.L.; Haney, E.F.; Bolscher, J.G.; Hyndman, M.E.; Hancock, R.E.; Vogel, H.J. Anticancer activities of bovine and human lactoferricin-derived peptides. Biochem. Cell Biol. 2017, 95, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Huertas, N.J.; Monroy, Z.J.R.; Medina, R.F.; Castañeda, J.E.G. Antimicrobial Activity of Truncated and Polyvalent Peptides Derived from the FKCRRWQWRMKKGLA Sequence against Escherichia coli ATCC 25922 and Staphylococcus aureus ATCC 25923. Molecules 2017, 22. [Google Scholar] [CrossRef] [PubMed]
- Huertas Méndez, N.J.; Vargas Casanova, Y.; Gómez Chimbi, A.K.; Hernández, E.; Leal Castro, A.L.; Melo Diaz, J.M.; Rivera Monroy, Z.J.; García Castañeda, J.E. Synthetic Peptides Derived from Bovine Lactoferricin Exhibit Antimicrobial Activity against E. coli ATCC 11775, S. maltophilia ATCC 13636 and S. enteritidis ATCC 13076. Molecules 2017, 22. [Google Scholar] [CrossRef] [PubMed]
- Solarte, V.; Rosas, J.E.; Rivera, Z.J.; Arango, M.L.; García, J.E.; Vernot, J.A. A Tetrameric Peptide Derived from Bovine Lactoferricin Exhibits Specific Cytotoxic Effects against Oral Squamous-Cell Carcinoma Cell Lines. Biomed. Res. Int. 2015, 2015, 630179. [Google Scholar] [CrossRef] [PubMed]
- Chapple, D.S.; Hussain, R.; Joannou, C.L.; Hancock, R.E.; Odell, E.; Evans, R.W. Structure and association of human lactoferrin peptides with Escherichia coli lipopolysaccharide. Antimicrob. Agents Chemother. 2004, 48, 2190–2198. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.F.; Jie, M.M.; Li, B.S.; Hu, C.J.; Xie, R.; Tang, B.; Yang, S.M. Peptide-Based Treatment: A Promising Cancer Therapy. J. Immunol. Res. 2015, 2015, 761820. [Google Scholar] [CrossRef] [PubMed]
- Solarte, V.A.; Conget, P.; Vernot, J.P.; Rosas, J.E.; Rivera, Z.J.; García, J.E.; Arango-Rodríguez, M.L. A tetrameric peptide derived from bovine lactoferricin as a potential therapeutic tool for oral squamous cell carcinoma: A preclinical model. PLoS ONE 2017, 12, e0174707. [Google Scholar] [CrossRef] [PubMed]
- Vergel, C.; Rivera, Z.J.; Rosas, J.E.; García, J.E. Efficient Synthesis of Peptides with 4-Methylpiperidine as Fmoc Removal Reagent by Solid Phase Synthesis. J. Mex. Chem. Soc. 2014, 58, 369–365. [Google Scholar]
- Douglas, S.; Hoskin, D.W.; Hilchie, A.L. Assessment of antimicrobial (host defense) peptides as anti-cancer agents. Methods Mol. Biol. 2014, 1088, 159–170. [Google Scholar] [PubMed]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds used in this paper are available from the authors. |





| Synthetic Peptide Sequence | Analytical Characterization | |||
|---|---|---|---|---|
| RP-HPLC | MALDI-TOF MS | |||
| tR (min) | Purity c (%) | Monoisotopic Mass (M) | Experimental m/z, [M + H]+ | |
| 20RRWQWR25 | 4.3 | 92 | 985.54 | 986.55 |
| (RRWQWR)2K-Ahx | 5.1 | 86 | 2195.24 | 2198.46 |
| (RRWQWR)2K-Ahx-C a | 5.2 | 86 | 2298.24 | 2300.93 |
| C-RRWQWR-Ahx-C b | 4.4 | 85 | 1304.64 | 1306.08 |
| 20RRWQWRMKKLG30 | 4.7 | 95 | 1542.87 | 1544.53 |
| (RRWQWRMKKLG)2K-Ahx | 5.3 | 86 | 3309.91 | 3311.62 |
| (RRWQWRMKKLG)2K-Ahx-C a | 5.4 | 90 | 3412.92 | 3415.07 |
| C-RRWQWRMKKLG-Ahx-C b | 5.0 | 95 | 1861.98 | 1862.14 |
| 17FKARRWQWRMKKLGA31 | 4.9 | 95 | 1960.11 | 1961.99 |
| (FKARRWQWRMKKLGA)2K-Ahx | 5.5 | 88 | 4144.38 | 4146.73 |
| (FKARRWQWRMKKLGA)2K-Ahx-C a | 5.5 | 85 | 4247.39 | 4255.51 |
| C-FKARRWQWRMKKLGA-Ahx-C b | 5.2 | 92 | 2279.21 | 2279.93 |
| Peptide Code | Antibacterial Effect MIC/MBC µg/mL (µM) | Cytotoxic Effect IC50 (µM) | ||
|---|---|---|---|---|
| E. coli ATCC 11775 | E. coli ATCC 25922 | MDA-MB-468 | MDA-MB-231 | |
| LfcinB (20–25) | 200(203)/200(203) | 200(203)/200(203) | >203 | >203 |
| LfcinB (20–25)2 | 50(22)/50(22) | 12.5(6)/25(11) | >100 | 130 |
| LfcinB (20–25)4 | 100(22)/200(44) | 100(22)/100(22) | 6 | 15 |
| LfcinB (20–25)cyc | 200(153)/200(153) | 100(77)/>200(>153) | >200 | >200 |
| LfcinB (20–30) | 200(130)/200(130) | 200(130)/200(130) | >130 | >130 |
| LfcinB (20–30)2 | 200(60)/200(60) | 100(30)/200(60) | 5 | 14 |
| LfcinB (20–30)4 | 200(15)/200(15) | 200(15)/200(15) | 2 | 6 |
| LfcinB (20–30)cyc | 200(107)/200(107) | 200(107)/200(107) | >107 | 27 |
| [Ala19]-LfcinB (17–31) | >200(>102)/>200(102) | 200(102)/200(102) | >102 | >102 |
| [Ala19]-LfcinB (17–31)2 | >200(>48)/>200(>48) | 100(24)/100(24) | 11 | 31 |
| [Ala19]-LfcinB (17–31)4 | ND | ND | 5 | 9 |
| [Ala19]-LfcinB (17–31)cyc | >200(>88)/>200(>88) | 200(88)/200(88) | >88 | >88 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vargas Casanova, Y.; Rodríguez Guerra, J.A.; Umaña Pérez, Y.A.; Leal Castro, A.L.; Almanzar Reina, G.; García Castañeda, J.E.; Rivera Monroy, Z.J. Antibacterial Synthetic Peptides Derived from Bovine Lactoferricin Exhibit Cytotoxic Effect against MDA-MB-468 and MDA-MB-231 Breast Cancer Cell Lines. Molecules 2017, 22, 1641. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules22101641
Vargas Casanova Y, Rodríguez Guerra JA, Umaña Pérez YA, Leal Castro AL, Almanzar Reina G, García Castañeda JE, Rivera Monroy ZJ. Antibacterial Synthetic Peptides Derived from Bovine Lactoferricin Exhibit Cytotoxic Effect against MDA-MB-468 and MDA-MB-231 Breast Cancer Cell Lines. Molecules. 2017; 22(10):1641. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules22101641
Chicago/Turabian StyleVargas Casanova, Yerly, Jorge Antonio Rodríguez Guerra, Yadi Adriana Umaña Pérez, Aura Lucía Leal Castro, Giovanni Almanzar Reina, Javier Eduardo García Castañeda, and Zuly Jenny Rivera Monroy. 2017. "Antibacterial Synthetic Peptides Derived from Bovine Lactoferricin Exhibit Cytotoxic Effect against MDA-MB-468 and MDA-MB-231 Breast Cancer Cell Lines" Molecules 22, no. 10: 1641. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules22101641