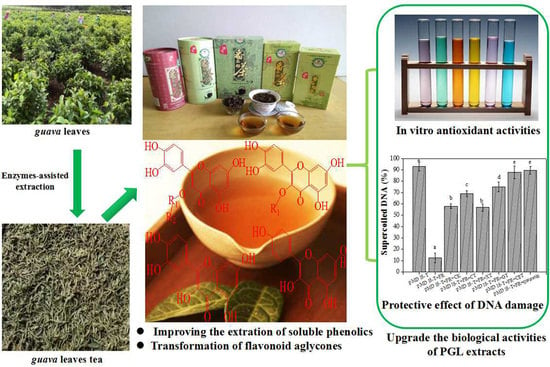
Complex Enzyme-Assisted Extraction Releases Antioxidative Phenolic Compositions from Guava Leaves
Abstract
:1. Introduction
2. Results
2.1. Changes of Total Phenolic and Flavonoid Contents with Enzyme-Assisted Extraction
2.2. Changes of Phenolic Compounds with Enzyme-Assisted Extraction
2.3. Changes of Bioactivity with Enzyme-Assisted Extraction
2.3.1. Antioxidant Activity
2.3.2. Protection Effect against DNA Damage
3. Discussion
3.1. Enzyme-Assisted Extraction Action on Total Soluble Phenolics and Soluble Flavonoids Contents
3.2. Enzyme-Assisted Extraction Action on the Phenolic Compositions
3.3. Enzyme-Assisted Extraction Action on Bioactivity
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Enzyme Pre-Treatment
4.3. Extraction of Free Phenolic Fractions
4.4. Extraction of Soluble-Conjugate Phenolics
4.5. Extraction of Insoluble-Bound Phenolics
4.6. Determination of Phenolic Content
4.7. Determination of Flavonoids Content
4.8. HPLC Analysis
4.9. Evaluation of Antioxidant Activity
4.9.1. ABTS Radical Cation (ABTS+) Scavenging Activity
4.9.2. DPPH Radical Scavenging Activity
4.9.3. Ferric Reducing Antioxidant Power (FRAP) Assay
4.10. Inhibition of Supercoiled DNA Strand Breakage
4.11. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
References
- Vadivel, V.; Biesalski, H.K. Contribution of phenolic compounds to the antioxidant potential and type II diabetes related enzyme inhibition properties of Pongamia pinnata L. Pierre seeds. Process Biochem. 2011, 46, 1973–1980. [Google Scholar] [CrossRef]
- Gutiérrez, R.M.P.; Mitchell, S.; Solis, R.V. Psidium guajava: A review of its traditional uses, phytochemistry and pharmacology. J. Ethnopharmacol. 2008, 117, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Bljajić, K.; Petlevski, R.; Vujić, L.; Čačić, A.; Šoštarić, N.; Jablan, J.; Zovko Končić, M. Chemical composition, antioxidant and α-glucosidase-inhibiting activities of the aqueous and hydroethanolic extracts of Vaccinium myrtillus leaves. Molecules 2017, 22, 703. [Google Scholar] [CrossRef] [PubMed]
- Ranilla, L.G.; Kwon, Y.I.; Apostolidis, E.; Shetty, K. Phenolic compounds, antioxidant activity and in vitro inhibitory potential against key enzymes relevant for hyperglycemia and hypertension of commonly used medicinal plants, herbs and spices in Latin America. Bioresour. Technol. 2010, 101, 4676–4689. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wei, W.; Tian, X.; Shi, K.; Wu, Z. Improving bioactivities of polyphenol extracts from Psidium guajava L. leaves through co-fermentation of Monascus anka GIM 3.592 and Saccharomyces cerevisiae GIM 2.139. Ind. Crop. Prod. 2016, 94, 206–215. [Google Scholar] [CrossRef]
- Ahmad, N.; Zuo, Y.; Lu, X.; Anwar, F.; Hameed, S. Characterization of free and conjugated phenolic compounds in fruits of selected wild plants. Food Chem. 2016, 190, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Yeo, J.; Shahidi, F. Effect of hydrothermal processing on changes of insoluble-bound phenolics of lentils. J. Funct. Foods 2017. [Google Scholar] [CrossRef]
- Madhujith, T.; Shahidi, F. Antioxidant potential of barley as affected by alkaline hydrolysis and release of insoluble-bound phenolics. Food Chem. 2009, 117, 615–620. [Google Scholar] [CrossRef]
- Adom, K.K.; Liu, R.H. Antioxidant activity of grains. J. Agric. Food Chem. 2002, 50, 6182–6187. [Google Scholar] [CrossRef] [PubMed]
- Naczk, M.; Shahidi, F. The effect of methanol-ammonia-water treatment on the content of phenolic acids of canola. Food Chem. 1989, 31, 159–164. [Google Scholar] [CrossRef]
- Li, F.; Mao, Y.D.; Wang, Y.F.; Raza, A.; Qiu, L.P.; Xu, X.Q. Optimization of ultrasonic-assisted enzymatic extraction conditions for improving total phenolic content, antioxidant and antitumor activities in vitro from Trapa quadrispinosa Roxb. residues. Molecules 2017, 22, 396. [Google Scholar] [CrossRef] [PubMed]
- Altemimi, A.; Choudhary, R.; Watson, D.G.; Lightfoot, D.A. Effects of ultrasonic treatments on the polyphenol and antioxidant content of spinach extracts. Ultrason. Sonochem. 2015, 24, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Dranca, F.; Oroian, M. Optimization of ultrasound-assisted extraction of total monomeric anthocyanin (TMA) and total phenolic content (TPC) from eggplant (Solanum melongena L.) peel. Ultrason. Sonochem. 2016, 31, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Vinatoru, M.; Mason, T.J.; Calinescu, I. Ultrasonically assisted extraction (UAE) and microwave assisted extraction (MAE) of functional compounds from plant materials. TrAC Trends Anal. Chem. 2017. [Google Scholar] [CrossRef]
- Wanyo, P.; Meeso, N.; Siriamornpun, S. Effects of different treatments on the antioxidant properties and phenolic compounds of rice bran and rice husk. Food Chem. 2014, 157, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Landbo, A.K.; Meyer, A.S. Enzyme-assisted extraction of antioxidative phenols from black currant juice press residues (Ribes nigrum). J. Agric. Food Chem. 2001, 49, 3169–3177. [Google Scholar] [CrossRef] [PubMed]
- Mathew, S.; Abraham, T.E. Ferulic acid: An antioxidant found naturally in plant cell walls and feruloyl esterases involved in its release and their applications. Crit. Rev. Biotechnol. 2004, 24, 59–83. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.Z.; Hwang, I.W.; Chung, S.K. Enhancing polyphenol extraction from unripe apples by carbohydrate-hydrolyzing enzymes. J. Zhejiang Univ. Sci. B 2009, 10, 912–919. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wen, W.; Zhang, R.; Wei, Z.; Deng, Y.; Xiao, J.; Zhang, M. Complex enzyme hydrolysis releases antioxidative phenolics from rice bran. Food Chem. 2017, 214, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Alshikh, N.; de Camargo, A.C.; Shahidi, F. Phenolics of selected lentil cultivars: Antioxidant activities and inhibition of low-density lipoprotein and DNA damage. J. Funct. Foods 2015, 18, 1022–1038. [Google Scholar] [CrossRef]
- Manach, C.; Morand, C.; Demigné, C.; Texier, O.; Régérat, F.; Rémésy, C. Bioavailability of rutin and quercetin in rats. FEBS Lett. 1997, 409, 12–16. [Google Scholar] [CrossRef]
- Wein, S.; Wolffram, S. Concomitant intake of quercetin with a grain-based diet acutely lowers postprandial plasma glucose and lipid concentrations in pigs. Biomed. Res. Int. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Wiczkowski, W.; Romaszko, J.; Bucinski, A.; Szawara-Nowak, D.; Honke, J.; Zielinski, H.; Piskula, M.K. Quercetin from shallots (Allium cepa L. var. aggregatum) is more bioavailable than its glucosides. J. Nutr. 2008, 138, 885–888. [Google Scholar] [PubMed]
- Boulila, A.; Hassen, I.; Haouari, L.; Mejri, F.; Amor, I.B.; Casabianca, H.; Hosni, K. Enzyme-assisted extraction of bioactive compounds from bay leaves (Laurus nobilis L.). Ind. Crop Prod. 2015, 74, 485–493. [Google Scholar] [CrossRef]
- Bhanja, T.; Kumari, A.; Banerjee, R. Enrichment of phenolics and free radical scavenging property of wheat koji prepared with two filamentous fungi. Bioresour. Technol. 2009, 100, 2861–2866. [Google Scholar] [CrossRef] [PubMed]
- Alrahmany, R.; Avis, T.J.; Tsopmo, A. Treatment of oat bran with carbohydrases increases soluble phenolic acid content and influences antioxidant and antimicrobial activities. Food Res. Int. 2013, 52, 568–574. [Google Scholar] [CrossRef]
- Shahidi, F.; Zhong, Y. Measurement of antioxidant activity. J. Funct. Foods 2015, 18, 757–781. [Google Scholar] [CrossRef]
- Ti, H.; Li, Q.; Zhang, R.; Zhang, M.; Deng, Y.; Wei, Z.; Zhang, Y. Free and bound phenolic profiles and antioxidant activity of milled fractions of different indica rice varieties cultivated in southern China. Food Chem. 2014, 159, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Alrahmany, R.; Tsopmo, A. Role of carbohydrases on the release of reducing sugar, total phenolics and on antioxidant properties of oat bran. Food Chem. 2012, 132, 413–418. [Google Scholar] [CrossRef] [PubMed]
- Hiramoto, K.; Ojima, N.; Sako, K.I.; Kikugawa, K. Effect of plant phenolics on the formation of the spin-adduct of hydroxyl radical and the DNA strand breaking by hydroxyl radical. Biol. Pharm. Bull. 1996, 19, 558–563. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Lemos, M.; Sharma, M.; Shriram, V. Antioxidant and DNA damage protecting activities of Eulophia nuda Lindl. Free Radic. Antioxid. 2013, 3, 55–60. [Google Scholar] [CrossRef]
- Zhang, L.; Yang, X.; Zhang, Y.; Wang, L.; Zhang, R. In vitro antioxidant properties of different parts of pomegranate flowers. Food Bioprod. Process. 2011, 89, 234–240. [Google Scholar] [CrossRef]
- Chandrasekara, A.; Shahidi, F. Antiproliferative potential and DNA scission inhibitory activity of phenolics from whole millet grains. J. Funct. Foods 2011, 3, 159–170. [Google Scholar] [CrossRef]
- Kim, J.K.; Kim, M.; Cho, S.G.; Kim, M.K.; Kim, S.W.; Lim, Y.H. Biotransformation of mulberroside A from Morus alba results in enhancement of tyrosinase inhibition. J. Ind. Microbiol. Biotechnol. 2010, 37, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.B.; Singh, B.N.; Singh, S.P.; Nautiyal, C.S. Solid-state cultivation of Trichoderma harzianum NBRI-1055 for modulating natural antioxidants in soybean seed matrix. Bioresour. Technol. 2010, 101, 6444–6453. [Google Scholar] [CrossRef] [PubMed]
- Bei, Q.; Liu, Y.; Wang, L.; Chen, G.; Wu, Z. Improving free, conjugated, and bound phenolic fractions in fermented oats (Avena sativa L.) with Monascus anka and their antioxidant activity. J. Funct. Foods 2017, 32, 185–194. [Google Scholar] [CrossRef]
- Kumar, G.S.; Nayaka, H.; Dharmesh, S.M.; Salimath, P.V. Free and bound phenolic antioxidants in amla (Emblica officinalis) and turmeric (Curcuma longa). J. Food Compos. Anal. 2006, 19, 446–452. [Google Scholar] [CrossRef]
- Wang, L.; Bei, Q.; Wu, Y.; Liao, W.; Wu, Z. Characterization of soluble and insoluble-bound polyphenols from Psidium guajava L. leaves co-fermented with Monascus anka and Bacillus sp. and their bio-activities. J. Funct. Foods 2017, 32, 149–159. [Google Scholar] [CrossRef]
- Cai, S.; Wang, O.; Wu, W.; Zhu, S.; Zhou, F.; Ji, B.; Cheng, Q. Comparative study of the effects of solid-state fermentation with three filamentous fungi on the total phenolics content (TPC), flavonoids, and antioxidant activities of subfractions from oats (Avena sativa L.). J. Agric. Food Chem. 2011, 60, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Sasipriya, G.; Siddhuraju, P. Effect of different processing methods on antioxidant activity of underutilized legumes, Entada scandens seed kernel and Canavalia gladiata seeds. Food Chem. Toxicol. 2012, 50, 2864–2872. [Google Scholar] [CrossRef] [PubMed]
- Hammi, K.M.; Jdey, A.; Abdelly, C.; Majdoub, H.; Ksouri, R. Optimization of ultrasound-assisted extraction of antioxidant compounds from Tunisian Zizyphus lotus fruits using response surface methodology. Food Chem. 2015, 184, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Liyana-Pathirana, C.M.; Shahidi, F. Importance of insoluble-bound phenolics to antioxidant properties of wheat. J. Agric. Food Chem. 2006, 54, 1256–1264. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of guava leaves are available from the authors. |



| Analytes | Stage | Free (mg/100 g DM) | Soluble-Conjugate (mg/100 g DM) | Insoluble-Bound (mg/100 g DM) | Total Soluble (mg/100 g DM) |
|---|---|---|---|---|---|
| Gallic acid | CK | 161.1 ± 1.11a | 118.3 ± 1.32a | 351.5 ± 2.63a | 279.5 ± 2.33a |
| CAE | 173.0 ± 1.31b | 131.4 ± 1.81b | 259.6 ± 1.53b | 304.4 ± 3.12b | |
| XAE | 163.4 ± 2.21a | 117.5 ± 2.12a | 348.8 ± 2.01a | 280.9 ± 4.33a | |
| GAE | 235.3 ± 1.90c | 145.7 ± 1.31c | 245.7 ± 1.41c | 381.1 ± 3.21c | |
| CEAE | 276.6 ± 2.62d | 162.2 ± 1.71d | 71.36 ± 1.45d | 438.8 ± 4.33d | |
| Chlorogenic acid | CK | 25.8 ± 0.31a | 26.3 ± 0.40a | N.D. | 52.1 ± 0.71a |
| CAE | 31.1 ± 0.25b | 29.4 ± 0.31b | N.D. | 60.5 ± 0.56b | |
| XAE | 23.7 ± 0.61b | 27.4 ± 1.22b | N.D. | 51.8 ± 1.83a | |
| GAE | 36.5 ± 0.37b | 30.7 ± 0.42b | N.D. | 67.2 ± 0.73b | |
| CEAE | 41.4 ± 0.22b | 30.3 ± 0.21a | N.D. | 71.7 ± 0.43a | |
| p-hydroxybenzoic acid | CK | 1.1 ± 0.05a | 2.6 ± 0.04b | 13.4 ± 0.1e | 3.66 ± 0.09b |
| CAE | 2.0 ± 0.01b | 2.2 ± 0.01c | 9.0 ± 0.1c | 4.2 ± 0.02c | |
| XAE | 1.0 ± 0.02a | 2.7 ± 0.02a | 11.8 ± 0.07d | 3.7 ± 0.04a | |
| GAE | 2.6 ± 0.05c | 3.1 ± 0.01d | 6.0 ± 0.01b | 5.7 ± 0.11d | |
| CEAE | 2.9 ± 0.03d | 4.0 ± 0.03e | 0.3 ± 0.01a | 6.9 ± 0.06e | |
| Caffeic acid | CK | N.D. | 4.9 ± 0.07a | 12.1 ± 0.11b | 4.9 ± 0.07a |
| CAE | N.D. | 5.8 ± 0.31a | 11.1 ± 0.05b | 5.8 ± 0.31a | |
| XAE | N.D. | 5.0 ± 0.07a | 14.5 ± 0.13b | 5.0 ± 0.07a | |
| GAE | N.D. | 8.3 ± 0.01b | 8.3 ± 0.07a | 8.3 ± 0.01b | |
| CEAE | N.D. | 9.1 ± 0.03b | 7.7 ± 0.05a | 9.1 ± 0.03b | |
| Rutin | CK | 1.8 ± 0.02a | N.D. | 5.8 ± 0.11d | 1.8 ± 0.02a |
| CAE | 2.0 ± 0.07b | N.D. | 3.5 ± 0.08b | 2.0 ± 0.07b | |
| XAE | 1.8 ± 0.04a | N.D. | 4.3 ± 0.02c | 1.8 ± 0.04a | |
| GAE | 0.5 ± 0.01b | N.D. | 3.3 ± 0.11b | 0.5 ± 0.01b | |
| CEAE | 0.2 ± 0.02c | N.D. | 2.7 ± 0.06a | 0.2 ± 0.02c | |
| p-Coumaric acid | CK | 15.8 ± 0.21a | 9.4 ± 0.29a | 136.8 ± 0.89d | 25.2 ± 0.50a |
| CAE | 21.4 ± 0.09a | 11.6 ± 0.17b | 124.4 ± 1.61c | 32.9 ± 0.31b | |
| XAE | 14.4 ± 0.07a | 9.3 ± 1.21a | 148.3 ± 2.35d | 23.7 ± 1.28a | |
| GAE | 27.7 ± 0.05b | 15.2 ± 0.67c | 82.7 ± 2.01b | 42.8 ± 1.01c | |
| CEAE | 29.6 ± 0.08b | 17.7 ± 0.18d | 34.9 ± 1.01a | 47.6 ± 0.63d | |
| Isoquercitrin | CK | 37.1 ± 0.43b | 26.5 ± 0.53a | 19.6 ± 0.23d | 63.6 ± 0.56a |
| CAE | 41.2 ± 0.91c | 27.7 ± 0.15b | 10.3 ± 1.01c | 68.9 ± 0.78b | |
| XAE | 35.5 ± 0.16a | 27.7 ± 0.45a | 12.3 ± 0.97e | 62.9 ± 0.08a | |
| GAE | 20.8 ± 0.34d | 31.2 ± 0.21b | 6.7 ± 0.26b | 52.0 ± 2.01c | |
| CEAE | 18.6 ± 0.79e | 32.3 ± 0.19b | 5.6 ± 0.31a | 51.0 ± 1.23d | |
| Sinapic acid | CK | 4.7 ± 0.09a | N.D. | 5.6 ± 0.23d | 4.7 ± 0.09a |
| CAE | 2.3 ± 0.03b | N.D. | 8.3 ± 1.01c | 2.3 ± 0.03b | |
| XAE | 2.1 ± 0.01a | N.D. | 6.2 ± 0.97e | 2.1 ± 0.01a | |
| GAE | 3.4 ± 0.02a | N.D. | 8.5 ± 0.26b | 3.4 ± 0.02a | |
| CEAE | 1.5 ± 0.02d | N.D. | 7.3 ± 0.31a | 1.5 ± 0.02d | |
| Ferulic acid | CK | N.D. | N.D. | 10.5 ± 0.19d | N.D. |
| CAE | N.D. | N.D. | 8.8 ± 0.13c | N.D. | |
| XAE | N.D. | N.D. | 9.9 ± 0.31c | N.D. | |
| GAE | N.D. | 5.6 ± 0.19a | 6.4 ± 0.08b | 5.6 ± 0.19a | |
| CEAE | N.D. | 7.5 ± 0.31b | 5.6 ± 0.11a | 7.5 ± 0.31b | |
| Quercetin-3-O-β-d-xylopyranoside | CK | 53.6 ± 1.21a | N.D. | N.D. | 53.6 ± 1.21a |
| CAE | 59.5 ± 0.61b | N.D. | N.D. | 59.5 ± 0.61b | |
| XAE | 48.2 ± 0.34a | N.D. | N.D. | 48.2 ± 0.34a | |
| GAE | 61.3 ± 0.48c | N.D. | N.D. | 61.3 ± 0.48c | |
| CEAE | 72.9 ± 1.05d | N.D. | N.D. | 72.9 ± 1.05d | |
| Quercetin-3-O-α-l-arabinoside | CK | 87.2 ± 2.67b | N.D. | 11.4 ± 0.41c | 87.2 ± 2.67b |
| CAE | 113.2 ± 3.01c | N.D. | 17.4 ± 0.26d | 113.2 ± 3.01c | |
| XAE | 89.6 ± 0.29b | N.D. | 10.5 ± 1.11c | 89.6 ± 0.29b | |
| GAE | 68.2 ± 2.31d | 15.1 ± 1.01b | 10.2 ± 0.07b | 83.2 ± 3.32d | |
| CEAE | 11.1 ± 1.61a | 4.2 ± 0.08a | 7.1 ± 0.03a | 15.3 ± 1.57a | |
| Avicularin | CK | 258.1 ± 1.79b | N.D. | 16.4 ± 0.11c | 258.1 ± 1.79b |
| CAE | 245.6 ± 1.94c | N.D. | 14.2 ± 0.14c | 245.6 ± 1.94c | |
| XAE | 249.4 ± 3.27b | N.D. | 15.7 ± 0.23c | 249.4 ± 3.27b | |
| GAE | 71.2 ± 2.29c | 1.0 ± 0.08a | 10.3 ± 0.31b | 72.3 ± 2.29c | |
| CEAE | 17.8 ± 0.21a | 1.2 ± 0.02a | 6.3 ± 0.05a | 18.9 ± 0.23a | |
| Quercitrin | CK | 107.1 ± 1.68b | N.D. | 11.3 ± 0.01b | 107.1 ± 1.68b |
| CAE | 118.1 ± 1.17c | N.D. | 9.6 ± 0.02b | 118.1 ± 1.17c | |
| XAE | 97.1 ± 1.45a | N.D. | 10.4 ± 0.03b | 97.1 ± 1.45a | |
| GAE | 123.8 ± 2.64d | N.D. | 5.2 ± 0.01a | 123.8 ± 2.64d | |
| CEAE | 112.9 ± 2.15b | 21.4 ± 0.37b | 4.9 ± 0.01a | 134.3 ± 2.52b | |
| Quercetin | CK | 34.8 ± 0.57a | 39.0 ± 1.61a | 113.1 ± 0.31e | 73.8 ± 2.14a |
| CAE | 59.5 ± 0.21b | 47.2 ± 1.00b | 98.4 ± 2.19c | 106.7 ± 1.21b | |
| XAE | 37.6 ± 1.21a | 35.2 ± 0.29a | 109.3 ± 1.45d | 72.8 ± 1.36a | |
| GAE | 134.1 ± 0.21c | 65.0 ± 1.15c | 25.4 ± 1.23b | 199.0 ± 1.36c | |
| CEAE | 177.0 ± 2.03d | 81.9 ± 1.29d | 10.3 ± 0.07a | 258.9 ± 3.32d | |
| Kaempferol | CK | 5.1 ± 0.02b | N.D. | N.D. | 5.1 ± 0.02b |
| CAE | 6.0 ± 0.07c | N.D. | N.D. | 6.0 ± 0.07c | |
| XAE | 5.0 ± 0.02a | N.D. | N.D. | 5.0 ± 0.02a | |
| GAE | 7.0 ± 0.03c | N.D. | N.D. | 7.0 ± 0.03c | |
| CEAE | 11.2 ± 0.01d | N.D. | N.D. | 11.2 ± 0.01d |
| Stage | Antioxidant Activity | |||
|---|---|---|---|---|
| Free | Soluble-Conjugate | Insoluble-Bound | Total Soluble | |
| ABTS value | (mmol TE/g DM) | |||
| CK | 20.6 ± 1.1Aa | 6.5 ± 0.4Ba | 20.2 ± 0.2Aa | 27.1 ± 1.4a |
| CAE | 23.5 ± 0.5Ab | 9.5 ± 1.6Cb | 16.8 ± 0.9Bb | 33.0 ± 2.0b |
| XAE | 18.7 ± 1.0Aa | 7.2 ± 0.4Ba | 20.6 ± 1.5Aa | 25.9 ± 1.4a |
| GAE | 29.0 ± 0.78Ac | 10.8 ± 0.4Bc | 12.2 ± 0.5Bc | 39.9 ± 1.1c |
| CEAE | 36.8 ± 0.4Ad | 17.7 ± 0.1Bd | 7.4 ± 1.2Cd | 55.5 ± 0.5d |
| DPPH value | (mmol TE/g DM) | |||
| CK | 15.4 ± 0.2Ba | 8.0 ± 0.1Ca | 21.3 ± 0.2Aa | 23.4 ± 0.3a |
| CAE | 17.6 ± 0.4Ab | 11.5 ± 0.3Bb | 18.2 ± 1.1Ab | 29.1 ± 0.7b |
| XAE | 15.5 ± 1.09Ba | 7.3 ± 0.9Ca | 22.5 ± 0.5Aa | 22.8 ± 1.9a |
| GAE | 26.2 ± 0.7Ac | 13.6 ± 1.1Cc | 18.4 ± 1.3Bc | 39.7 ± 1.8c |
| CEAE | 35.1 ± 0.04Ad | 17.9 ± 1.2Bd | 11.1 ± 1.0Cd | 53.0 ± 1.3d |
| FARP value | (μmol Fe(II)SE/g DM) | |||
| CK | 83.7 ± 2.0Aa | 43.6 ± 1.0Ca | 56.5 ± 1.4Ba | 127.3 ± 5.5a |
| CAE | 94.8 ± 4.2Ab | 49.7 ± 2.3Bb | 50.5 ± 2.8Bb | 144.4 ± 3.8b |
| XAE | 79.5 ± 1.37Aa | 41.6 ± 3.4Ca | 54.2 ± 1.4Ba | 121.0 ± 6.5a |
| GAE | 111.8 ± 4.7Ac | 60.0 ± 2.1Bc | 37.9 ± 0.9Cc | 171.8 ± 6.1c |
| CEAE | 162.6 ± 2.5Ad | 79.7 ± 3.8Bd | 22.7 ± 1.3Cd | 242.3 ± 7.6d |
| Analytes | Regression Equation a | R2 | LOD b (μg/mL) | LOQ b (μg/mL) | Linear Range (μg/mL) | Recovery Rate (%, n = 4) |
|---|---|---|---|---|---|---|
| Gallic acid | Y = 3.56 × 107X + 2.06 × 104 | 0.9921 | 0.024 | 0.076 | 5.64–70.5 | 98.99 |
| Chlorogenic acid | Y = 2.51 × 107X + 1.93 × 104 | 0.9931 | 0.046 | 0.065 | 0.5–50 | 99.13 |
| p-hydroxybenzoic acid | Y = 1.67 × 107X + 3.11 × 104 | 0.9978 | 0.078 | 0.073 | 0.5–50 | 97.98 |
| Caffeic acid | Y = 8.53 × 106X + 1.48 × 104 | 0.9989 | 0.009 | 0.021 | 0.5–50 | 100.01 |
| Rutin | Y = 1.99 × 107X + 5.37 × 104 | 0.9989 | 0.019 | 0.053 | 6–75 | 99.02 |
| p-Coumaric acid | Y = 6.97 × 107X + 1.13 × 105 | 0.9981 | 0.031 | 0.024 | 0.5–50 | 98.99 |
| Isoquercitrin | Y = 1.24 × 107X + 1.40 × 103 | 0.9992 | 0.067 | 0.048 | 6–75 | 100.13 |
| Sinapic acid | Y = 1.87 × 107X + 2.93 × 104 | 0.9991 | 0.078 | 0.017 | 0.5–50 | 95.31 |
| Ferulic acid | Y = 4.68 × 107X + 6.91 × 104 | 0.9978 | 0.065 | 0.029 | 0.5–50 | 99.89 |
| Quercetin-3-O-β-d-xylopyranoside | Y = 1.78 × 107X + 1.40 × 104 | 0.9989 | 0.039 | 0.054 | 6–75 | 99.78 |
| Quercetin-3-O-α-l-arabinoside | Y = 1.48 × 107X + 2.03 × 103 | 0.9996 | 0.048 | 0.051 | 6–75 | 98.97 |
| Avicularin | Y = 1.45 × 107X + 1.96 × 103 | 0.9997 | 0.051 | 0.023 | 6–75 | 99.86 |
| Quercitrin | Y = 1.30 × 107X + 5.98 × 103 | 0.9978 | 0.024 | 0.018 | 6–75 | 99.13 |
| Quercetin | Y = 1.74 × 107X − 1.45 × 104 | 0.9992 | 0.031 | 0.010 | 2–75 | 101.07 |
| Kaempferol | Y = 8.83 × 107X − 2.23 × 104 | 0.9992 | 0.076 | 0.037 | 6–75 | 99.65 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Wu, Y.; Liu, Y.; Wu, Z. Complex Enzyme-Assisted Extraction Releases Antioxidative Phenolic Compositions from Guava Leaves. Molecules 2017, 22, 1648. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules22101648
Wang L, Wu Y, Liu Y, Wu Z. Complex Enzyme-Assisted Extraction Releases Antioxidative Phenolic Compositions from Guava Leaves. Molecules. 2017; 22(10):1648. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules22101648
Chicago/Turabian StyleWang, Lu, Yanan Wu, Yan Liu, and Zhenqiang Wu. 2017. "Complex Enzyme-Assisted Extraction Releases Antioxidative Phenolic Compositions from Guava Leaves" Molecules 22, no. 10: 1648. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules22101648