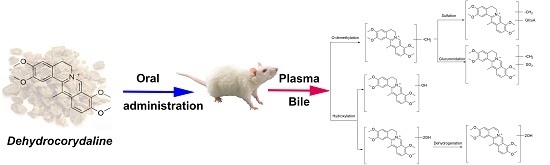
Identification of Metabolites of the Cardioprotective Alkaloid Dehydrocorydaline in Rat Plasma and Bile by Liquid Chromatography Coupled with Triple Quadrupole Linear Ion Trap Mass Spectrometry
Abstract
:1. Introduction
2. Results and Discussion
2.1. Optimization of HPLC Conditions
2.2. Optimization of Sample Preparation
2.3. Identification of DHC
2.4. Identification of Metabolites
2.5. Structure Elucidation of DHC Metabolites
2.5.1. Metabolites Produced by O-demethylation (M1, M2, M3 and M4)
2.5.2. Metabolites Produced by Glucuronidation of O-Demethyl-DHC (M5, M6 and M7)
2.5.3. Metabolites Produced by Sulfation of O-demethyl-DHC (M8, M9 and M10)
2.5.4. Metabolites Produced by Hydroxylation (M11, M12, M13 and M14)
2.5.5. Metabolites Produced by Di-Hydroxylation (M15 and M16)
2.5.6. Metabolites Produced by Di-Hydroxylation and Dehydrogenation (M17 and M18)
2.6. A Summary of Metabolic Pathways for DHC
3. Experimental Section
3.1. Materials and Reagents
3.2. Instruments and LC–MS/MS Conditions
3.3. Animal Handling
3.4. Sample Preparation
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ling, H.; Wu, L.; Li, L. Corydalis yanhusuo rhizoma extract reduces infarct size and improves heart function during myocardial ischemia/reperfusion by inhibiting apoptosis in rats. Phytother. Res. PTR 2006, 20, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.N.; Zhai, L.; Zhang, Q.M.; Jiang, M.X.; Ye, W.; Wang, R.W. To observe the clinical efficacy and safety on treating coronary heart disease and angina pectoris with kodaling tablets. Clin. J. Chin. Med. 2014, 6, 31–34. [Google Scholar]
- Xu, Z.; Chen, X.; Fu, S.; Bao, J.; Dang, Y.; Huang, M.; Chen, L.; Wang, Y. Dehydrocorydaline inhibits breast cancer cells proliferation by inducing apoptosis in mcf-7 cells. Am. J. Chin. Med. 2012, 40, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Grycova, L.; Dostal, J.; Marek, R. Quaternary protoberberine alkaloids. Phytochemistry 2007, 68, 150–175. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.R.; Wu, Q.X.; Shi, H.L.; Chen, W.P.; Chang, S.Q.; Zhao, S.Y.; Tian, X.Y.; Zhou, L.F.; Guo, S.M.; Li, Y.J. Pharmacological actions of dehydrocorydaline on cardiovascular system. Acta Pharm. Sin. 1982, 17, 61–65. [Google Scholar]
- Li, Q.; Guan, H.; Wang, X.; He, Y.; Sun, H.; Tan, W.; Luo, X.; Su, M.; Shi, Y. Fingerprint-efficacy study of the quaternary alkaloids in corydalis yanhusuo. J. Ethnopharmacol. 2017, 207, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Li, C.; Liu, X. Determination of dehydrocorydaline content in kedaling tablets by hplc. J. Hunan Univ. Chin. Med. 2012, 32, 37–39. [Google Scholar]
- Fujii, T.; Miyazaki, H.; Nambu, K.; Kagemoto, A.; Hashimoto, M. Disposition and metabolism of 14c-dehydrocorydaline in mice and rats. Radioisotopes 1984, 33, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Xing, D.; Jin, W.; Sun, H.; Shi, Y. Chemical fingerprint and metabolic fingerprint analysis of the medicinal extract of corydalis yanhusuo by hplc-uv and hplc-ms methods. Asian J. Chem. 2011, 23, 723–728. [Google Scholar]
- Li, Q.Y.; Li, K.T.; Sun, H.; Jin, W.; Shi, J.W.; Shi, Y. Lc-ms/ms determination and pharmacokinetic study of dehydrocorydaline in rat plasma after oral administration of dehydrocorydaline and corydalis yanhusuo extract. Molecules 2014, 19, 16312–16326. [Google Scholar] [CrossRef] [PubMed]
- Vrba, J.; Papouskova, B.; Pyszkova, M.; Zatloukalova, M.; Lemr, K.; Ulrichova, J.; Vacek, J. Metabolism of palmatine by human hepatocytes and recombinant cytochromes p450. J. Pharm. Biomed. Anal. 2015, 102, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Liu, Z.; Guo, M.; Liu, S. Structural elucidation and identification of alkaloids in rhizoma coptidis by electrospray ionization tandem mass spectrometry. J. Mass Spectrom. 2004, 39, 1356–1365. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, H.; Si, N.; Ren, W.; Han, L.; Xin, S.; Zuo, R.; Wei, X.; Yang, J.; Zhao, H.; et al. Metabolic profiling analysis of berberine, palmatine, jatrorrhizine, coptisine and epiberberine in zebrafish by ultra-high performance liquid chromatography coupled with ltq orbitrap mass spectrometer. Xenobiot. Fate Foreign Compd. Biol. Syst. 2015, 45, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.C.; Wu, W.H.; Han, F.M.; Chen, Y. Identification of in vivo and in vitro metabolites of palmatine by liquid chromatography-tandem mass spectrometry. J. Pharm. Pharmacol. 2009, 61, 647–652. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Han, F.; Chen, H.; Peng, Z.; Chen, Y. Identification of palmatine and its metabolites in rat urine by liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. RCM 2007, 21, 2019–2022. [Google Scholar] [CrossRef] [PubMed]
- Matsukawa, N.; Matsumoto, M.; Hara, H. High biliary excretion levels of quercetin metabolites after administration of a quercetin glycoside in conscious bile duct cannulated rats. Biosci. Biotechnol. Biochem. 2009, 73, 1863–1865. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |









| No. | Parent Ion m/z | Retention Time (min) | Formula | Product Ions (MS2) |
|---|---|---|---|---|
| DHC | 366 | 28.4 | C22H24NO4+ | 351, 350, 334, 322, 308, 290, 278 |
| M1, M2, M3, M4 | 352 | 15.0, 15.9, 16.7, 19.5 | C21H22NO4+ | 337, 336, 322, 309, 308, 293 |
| M5, M6, M7 | 528 | 4.4, 5.0, 5.5 | C27H30NO10+ | 352, 337, 322 308, 293 |
| M8, M9, M10 | 432 | 10.9, 12.4, 16.0 | C21H22NSO7+ | 352, 337, 322, 308, 293 |
| M11 | 382 | 7.2 | C22H24NO5+ | 367, 366, 364, 352, 350, 338, 324, 321, 308, 305, 294 |
| M12, M13, M14 | 382 | 11.3, 15.7, 27.8 | C22H24NO5+ | 367, 366, 352, 350, 349, 338, 324, 321, 306 |
| M15 | 398 | 12.3 | C22H24NO6+ | 383, 382, 368, 354, 340, 338, 324, 322, 294, 266 |
| M16 | 398 | 13.8 | C22H24NO6+ | 383, 382, 380, 368, 366, 365, 354, 340, 337 |
| M17 | 396 | 4.7 | C22H22NO6+ | 381, 380, 378, 366, 364, 352, 350, 338, 335, 322 |
| M18 | 396 | 7.7 | C22H22NO6+ | 381, 380, 366, 364, 352, 350, 337, 336, 332 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guan, H.; Li, K.; Wang, X.; Luo, X.; Su, M.; Tan, W.; Chang, X.; Shi, Y. Identification of Metabolites of the Cardioprotective Alkaloid Dehydrocorydaline in Rat Plasma and Bile by Liquid Chromatography Coupled with Triple Quadrupole Linear Ion Trap Mass Spectrometry. Molecules 2017, 22, 1686. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules22101686
Guan H, Li K, Wang X, Luo X, Su M, Tan W, Chang X, Shi Y. Identification of Metabolites of the Cardioprotective Alkaloid Dehydrocorydaline in Rat Plasma and Bile by Liquid Chromatography Coupled with Triple Quadrupole Linear Ion Trap Mass Spectrometry. Molecules. 2017; 22(10):1686. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules22101686
Chicago/Turabian StyleGuan, Huanyu, Kaitong Li, Xiaoming Wang, Xiaomei Luo, Meifeng Su, Wenting Tan, Xiaoyan Chang, and Yue Shi. 2017. "Identification of Metabolites of the Cardioprotective Alkaloid Dehydrocorydaline in Rat Plasma and Bile by Liquid Chromatography Coupled with Triple Quadrupole Linear Ion Trap Mass Spectrometry" Molecules 22, no. 10: 1686. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules22101686