Cinnamic Acid Analogs as Intervention Catalysts for Overcoming Antifungal Tolerance
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of the Most Potent Cinnamic Acids via Yeast Screening: 4-Chloro-α-methyl-, 4-Methoxy-, 4-Methyl- and 3-Methylcinnamic Acids
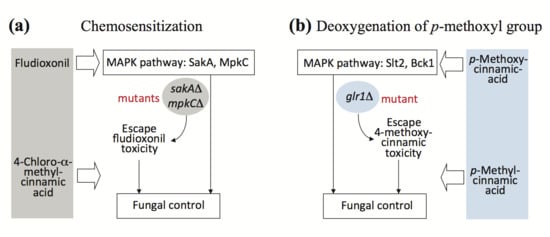
2.2. Fungal Tolerance to 4-Methoxycinnamic Acid Was Unique to Glutathione Reductase Mutant
2.3. Supplementation of Reduced Glutathione (GSH) Did Not Abolish Hyper-Tolerance to 4-Methoxycinnamic Acid
2.4. Overcoming Fludioxonil Tolerance of Aspergillus Fumigatus Antioxidant MAPK Mutants by 4-Chloro-α-methylcinnamic Acid
2.5. Antifungal Chemosensitization of 4-Chloro-α-methyl- or 4-Methylcinnamic Acid to CAS or OG
3. Materials and Methods
3.1. Fungal Strains and Culture Conditions
3.2. Antifungal Susceptibility Testing
3.2.1. Yeast Dilution Bioassay in Saccharomyces cerevisiae
3.2.2. Agar Plate Bioassay in Aspergillus fumigatus: Susceptibility of WT and MAPK Mutants to Cinnamic Acid Derivatives
3.2.3. Liquid Bioassay in Aspergillus brasiliensis (CLSI Protocol)
3.3. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Latge, J.P.; Beauvais, A.; Chamilos, G. The cell wall of the human fungal pathogen Aspergillus fumigatus: Biosynthesis, organization, immune response, and virulence. Annu. Rev. Microbiol. 2017, 71, 99–116. [Google Scholar] [CrossRef] [PubMed]
- Perlin, D.S. Mechanisms of echinocandin antifungal drug resistance. Ann. N. Y. Acad. Sci. 2015, 1354, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Levin, D.E.; Fields, F.O.; Kunisawa, R.; Bishop, J.M.; Thorner, J. A candidate protein kinase C gene, PKC1, is required for the S. cerevisiae cell cycle. Cell 1990, 62, 213–224. [Google Scholar] [CrossRef]
- Fuchs, B.B.; Mylonakis, E. Our paths might cross: The role of the fungal cell wall integrity pathway in stress response and cross talk with other stress response pathways. Eukaryot. Cell 2009, 8, 1616–1625. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, T.; Mizutani, O.; Furukawa, K.; Sato, N.; Yoshimi, A.; Yamagata, Y.; Nakajima, T.; Abe, K. MpkA-dependent and -independent cell wall integrity signaling in Aspergillus nidulans. Eukaryot. Cell 2007, 6, 1497–1510. [Google Scholar] [CrossRef] [PubMed]
- García-Rodriguez, L.J.; Durán, A.; Roncero, C. Calcofluor antifungal action depends on chitin and a functional high-osmolarity glycerol response (HOG) pathway: Evidence for a physiological role of the Saccharomyces cerevisiae HOG pathway under noninducing conditions. J. Bacteriol. 2000, 182, 2428–2437. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Ram, A.F.; Sheraton, J.; Klis, F.M.; Bussey, H. Regulation of cell wall beta-glucan assembly: PTC1 negatively affects PBS2 action in a pathway that includes modulation of EXG1 transcription. Mol. Gen. Genet. 1995, 248, 260–269. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.H.; Silverman, S.J.; Gaughran, J.P.; Kirsch, D.R. Multiple copies of PBS2, MHP1 or LRE1 produce glucanase resistance and other cell wall effects in Saccharomyces cerevisiae. Yeast 1997, 13, 199–213. [Google Scholar] [CrossRef]
- Alonso-Monge, R.; Navarro-García, F.; Molero, G.; Diez-Orejas, R.; Gustin, M.; Pla, J.; Sánchez, M.; Nombela, C. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J. Bacteriol. 1999, 181, 3058–3068. [Google Scholar] [PubMed]
- Altwasser, R.; Baldin, C.; Weber, J.; Guthke, R.; Kniemeyer, O.; Brakhage, A.A.; Linde, J.; Valiante, V. Network modeling reveals cross talk of MAP kinases during adaptation to caspofungin stress in Aspergillus fumigatus. PLoS ONE 2015, 10, e0136932. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Peña, J.M.; García, R.; Nombela, C.; Arroyo, J. The high-osmolarity glycerol (HOG) and cell wall integrity (CWI) signalling pathways interplay: A yeast dialogue between MAPK routes. Yeast 2010, 27, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Campbell, B.C.; Chan, K.L.; Kim, J.H. Chemosensitization as a means to augment commercial antifungal agents. Front. Microbiol. 2012, 3, 79. [Google Scholar] [CrossRef] [PubMed]
- Youngsaye, W.; Hartland, C.L.; Morgan, B.J.; Ting, A.; Nag, P.P.; Vincent, B.; Mosher, C.A.; Bittker, J.A.; Dandapani, S.; Palmer, M.; et al. ML212: A small-molecule probe for investigating fluconazole resistance mechanisms in Candida albicans. Beilstein J. Org. Chem. 2013, 9, 1501–1507. [Google Scholar] [CrossRef] [PubMed]
- Keniya, M.V.; Fleischer, E.; Klinger, A.; Cannon, R.D.; Monk, B.C. Inhibitors of the Candida albicans major facilitator superfamily transporter Mdr1p responsible for fluconazole resistance. PLoS ONE 2015, 10, e0126350. [Google Scholar] [CrossRef] [PubMed]
- Shirazi, F.; Kontoyiannis, D.P. Mitochondrial respiratory pathways inhibition in Rhizopus oryzae potentiates activity of posaconazole and itraconazole via apoptosis. PLoS ONE 2013, 8, e63393. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food and Drug Administration (FDA). Everything Added to Food in the United States. 2011. Available online: http://www.fda.gov/Food/IngredientsPackagingLabeling/FoodAdditivesIngredients/ucm115326.htm (accessed on 25 August 2017).
- Fungicide Resistance Action Committee. Available online: http://www.frac.info (accessed on 24 August 2017).
- Ma, C.M.; Abe, T.; Komiyama, T.; Wang, W.; Hattori, M.; Daneshtalab, M. Synthesis, anti-fungal and 1,3-beta-d-glucan synthase inhibitory activities of caffeic and quinic acid derivatives. Bioorg. Med. Chem. 2010, 18, 7009–7014. [Google Scholar] [CrossRef] [PubMed]
- Reinoso-Martín, C.; Schüller, C.; Schuetzer-Muehlbauer, M.; Kuchler, K. The yeast protein kinase C cell integrity pathway mediates tolerance to the antifungal drug caspofungin through activation of Slt2p mitogen-activated protein kinase signaling. Eukaryot. Cell 2003, 2, 1200–1210. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.; Rowan, R.; McCann, M.; Kavanagh, K. Exposure to caspofungin activates Cap and Hog pathways in Candida albicans. Med. Mycol. 2009, 47, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Couto, N.; Wood, J.; Barber, J. The role of glutathione reductase and related enzymes on cellular redox homoeostasis network. Free Radic. Biol. Med. 2016, 95, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Saccharomyces Genome Database. Available online: http://www.yeastgenome.org (accessed on 24 August 2017).
- Adisakwattana, S.; Hsu, W.H.; Yibchok-anun, S. Mechanisms of p-methoxycinnamic acid-induced increase in insulin secretion. Horm. Metab. Res. 2011, 43, 766–773. [Google Scholar] [CrossRef] [PubMed]
- Gunasekaran, S.; Venkatachalam, K.; Jeyavel, K.; Namasivayam, N. Protective effect of p-methoxycinnamic acid, an active phenolic acid against 1,2-dimethylhydrazine-induced colon carcinogenesis: Modulating biotransforming bacterial enzymes and xenobiotic metabolizing enzymes. Mol. Cell Biochem. 2014, 394, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Chamkha, M.; Garcia, J.L.; Labat, M. Metabolism of cinnamic acids by some Clostridiales and emendation of the descriptions of Clostridium aerotolerans, Clostridium celerecrescens and Clostridium xylanolyticum. Int. J. Syst. Evol. Microbiol. 2001, 51, 2105–2111. [Google Scholar] [CrossRef] [PubMed]
- Aldred, K.J.; Blower, T.R.; Kerns, R.J.; Berger, J.M.; Osheroff, N. Fluoroquinolone interactions with Mycobacterium tuberculosis gyrase: Enhancing drug activity against wild-type and resistant gyrase. Proc. Natl. Acad. Sci. USA 2016, 113, E839–E846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kojima, K.; Takano, Y.; Yoshimi, A.; Tanaka, C.; Kikuchi, T.; Okuno, T. Fungicide activity through activation of a fungal signalling pathway. Mol. Microbiol. 2004, 53, 1785–1796. [Google Scholar] [CrossRef] [PubMed]
- Reyes, G.; Romans, A.; Nguyen, C.K.; May, G.S. Novel mitogen-activated protein kinase MpkC of Aspergillus fumigatus is required for utilization of polyalcohol sugars. Eukaryot. Cell 2006, 5, 1934–1940. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.; Nguyen, C.K.; Romans, A.; May, G.S. A mitogen-activated protein kinase that senses nitrogen regulates conidial germination and growth in Aspergillus fumigatus. Eukaryot. Cell 2004, 3, 557–560. [Google Scholar] [CrossRef] [PubMed]
- Guillen, F.; Evans, C.S. Anisaldehyde and veratraldehyde acting as redox cycling agents for H2O2 production by Pleurotus eryngii. Appl. Environ. Microbiol. 1994, 60, 2811–2817. [Google Scholar] [PubMed]
- Jacob, C. A scent of therapy: Pharmacological implications of natural products containing redox-active sulfur atoms. Nat. Prod. Rep. 2006, 23, 851–863. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi: Approved Standard–Second Edition. CLSI Document M38-A2; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008; Volume 28. [Google Scholar]
- Moser, C.L.; Meyer, B.K. Comparison of compendial antimicrobial effectiveness tests: A review. AAPS PharmSciTech 2011, 12, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.A.; Lee, K.K.; Munro, C.A.; Gow, N.A. Caspofungin treatment of Aspergillus fumigatus results in ChsG-dependent upregulation of chitin synthesis and the formation of chitin-rich micro-colonies. Antimicrob. Agents Chemother. 2015. [Google Scholar] [CrossRef] [PubMed]
- Buckley, H.L.; Hart-Cooper, W.M.; Kim, J.H.; Faulkner, D.M.; Cheng, L.W.; Chan, K.L.; Vulpe, C.D.; Orts, W.J.; Amrose, S.E.; Mulvihill, M.J. Design and testing of safer, more effective preservatives for consumer products. ACS Sustain. Chem. Eng. 2017, 5, 4320–4331. [Google Scholar] [CrossRef]
- Kim, J.H.; Mahoney, N.; Chan, K.L.; Campbell, B.C.; Haff, R.P.; Stanker, L.H. Use of benzo analogs to enhance antimycotic activity of kresoxim methyl for control of aflatoxigenic fungal pathogens. Front. Microbiol. 2014, 5, 87. [Google Scholar] [CrossRef] [PubMed]
- Bisogno, F.; Mascoti, L.; Sanchez, C.; Garibotto, F.; Giannini, F.; Kurina-Sanz, M.; Enriz, R. Structure-antifungal activity relationship of cinnamic acid derivatives. J. Agric. Food Chem. 2007, 55, 10635–10640. [Google Scholar] [CrossRef] [PubMed]
- Tawata, S.; Taira, S.; Kobamoto, N.; Zhu, J.; Ishihara, M.; Toyama, S. Synthesis and antifungal activity of cinnamic acid esters. Biosci. Biotechnol. Biochem. 1996, 60, 909–910. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Chen, D.; Li, B.; Zhang, B.; Miao, F.; Zhou, L. Bioactivity and structure-activity relationship of cinnamic acid esters and their derivatives as potential antifungal agents for plant protection. PLoS ONE 2017, 12, e0176189. [Google Scholar] [CrossRef] [PubMed]
- Odds, F.C. Synergy, antagonism, and what the chequerboard puts between them. J. Antimicrob. Chemother. 2003, 52, 1. [Google Scholar] [CrossRef] [PubMed]
- Kirkman, T.W. Statistics to Use. Available online: http://www.physics.csbsju.edu/stats/ (accessed on 24 August 2017).
Sample Availability: Samples of the compounds, viz., all cinnamic acid derivatives, are available from the authors or vendors. |





| Cinnamic Acids | WT | slt2Δ | bck1Δ | glr1Δ |
|---|---|---|---|---|
| Group 1 (Highest activity): 4-Chloro-α-methylcinnamic acid | 0 | 0 | 0 | 0 |
| 4-Methylcinnamic acid | 1 | 0 | 0 | 0 |
| 4-Methoxycinnamic acid | 1 | 0 | 0 | 6 |
| 3-Methylcinnamic acid | 1 | 0 | 0 | 6 |
| Group 2 (Moderate activity): 2-Chloro-α-methylcinnamic acid | 6 | 2 | 2 | 4 |
| α-Methylcinnamic acid | 6 | 4 | 6 | 6 |
| 3,4-Dimethoxycinnamic acid | 6 | 4 | 6 | 6 |
| 4-Ethoxy-3-methoxycinnamic acid | 6 | 4 | 6 | 6 |
| Benzylcinnamic acid | 6 | 5 | 5 | 6 |
| 3,4-Dihydroxy-trans-cinnamic acid | 6 | 5 | 5 | 6 |
| 3-Chloro-4-methoxycinnamic acid | 6 | 5 | 6 | 6 |
| β-Methylcinnamic acid | 6 | 5 | 6 | 6 |
| Group 3 (No activity): Cinnamic acid (Basic structure) | 6 | 6 | 6 | 6 |
| Methyl-trans-cinnamic acid | 6 | 6 | 6 | 6 |
| 2-Methylcinnamic acid | 6 | 6 | 6 | 6 |
| 2-Methoxycinnamic acid | 6 | 6 | 6 | 6 |
| 3-Methoxycinnamic acid | 6 | 6 | 6 | 6 |
| 3,4,5-Trimethoxycinnamic acid | 6 | 6 | 6 | 6 |
| 4-Hydroxy-3-methoxycinnamic acid | 6 | 6 | 6 | 6 |
| 3-Hydroxy-4-methoxycinnamic acid | 6 | 6 | 6 | 6 |
| 2-Hydroxycinnamic acid | 6 | 6 | 6 | 6 |
| 3-Hydroxycinnamic acid | 6 | 6 | 6 | 6 |
| 4-Hydroxycinnamic acid | 6 | 6 | 6 | 6 |
| Cinnamyl acetate | 6 | 6 | 6 | 6 |
| 4-Acetoxy-3-methoxycinnamic acid | 6 | 6 | 6 | 6 |
| 4-Hexadecyloxy-3-methoxycinnamic acid | 6 | 6 | 6 | 6 |
| 2-Methoxy-α-methylcinnamic acid | 6 | 6 | 6 | 6 |
| Methyl α-methylcinnamic acid | 6 | 6 | 6 | 6 |
| Ethyl 4-hydroxy-3-methoxycinnamic acid | 6 | 6 | 6 | 6 |
| 4-Benzyloxy-3-methoxycinnamic acid | 6 | 6 | 6 | 6 |
| 3,5-Dimethoxy-4-hydroxycinnamic acid | 6 | 6 | 6 | 6 |
| 4-Amino-2-methylcinnamic acid | 6 | 6 | 6 | 6 |
| 3,4-Dihydroxyhydrocinnamic acid | 6 | 6 | 6 | 6 |
| 3-Hydroxy-α-mercapto-β-methylcinnamic acid | 6 | 6 | 6 | 6 |
| S. cerevisiae | Functions of Deleted Genes | 4-Methoxy- | 4-Methyl- |
|---|---|---|---|
| WT | Parental (Mat a his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) | 1 | 1 |
| glr1Δ | GLR1 & GLR1 interacting genes: Glutathione reductase | 4 (Hyper-tolerant) | 0 |
| trr1Δ | Cytoplasmic thioredoxin reductase | 0 | 0 |
| trr2Δ | Mitochondrial thioredoxin reductase | 1 | 0 |
| tsa1Δ | Thioredoxin peroxidase | 0 | 0 |
| trx1Δ | Cytoplasmic thioredoxin isoenzyme | 0 | 0 |
| trx2Δ | Cytoplasmic thioredoxin isoenzyme | 0 | 0 |
| gsh1Δ | γ-glutamylcysteine synthetase | 0 | 0 |
| gsh2Δ | Glutathione synthetase | 0 | 0 |
| grx1Δ | Glutathione-dependent disulfide oxidoreductase | 1 | 0 |
| grx2Δ | Cytoplasmic glutaredoxin | 0 | 0 |
| ahp1Δ | Thiol-specific peroxiredoxin | 1 | 0 |
| prx1Δ | Mitochondrial peroxiredoxin | 0 | 0 |
| tsa2Δ | Cytoplasmic thioredoxin peroxidase | 0 | 0 |
| dot5Δ | Nuclear thiol peroxidase | 0 | 0 |
| ycf1Δ | Vacuolar glutathione S-conjugate transporter | 1 | 0 |
| ctt1Δ | CAS responsive antioxidant genes: Cytosolic catalase T | 0 | 0 |
| cta1Δ | Catalase A | 0 | 0 |
| sod1Δ | Cytosolic Cu,Zn superoxide dismutase | 0 | 0 |
| sod2Δ | Mitochondrial Mn superoxide dismutase | 0 | 0 |
| Compounds | MIC Alone | MIC Combined | FICI | MFC Alone | MFC Combined | FFCI |
|---|---|---|---|---|---|---|
| 4-Chloro-α-methyl | 3.2 | 1.6 | 1 | 12.8 | 12.8 | 2 |
| CAS | 32 | 16 | 32 | 32 | ||
| 4-Methyl | 12.8 | 6.4 | 0.8 | 12.8 | 12.8 | 2 |
| CAS | 32 | 8 | 32 | 32 | ||
| 4-Methoxy | 12.8 | 12.8 | 2 | 12.8 | 12.8 | 2 |
| CAS | 32 | 32 | 32 | 32 | ||
| Mean: Chemosensitizers | 9.6 | 6.9 | − | 12.8 | 12.8 | − |
| CAS | 32 | 18.7 | − | 32 | 32 | − |
| t-test 1: Chemosensitizers | − | p = 0.590 | − | − | p = 1.000 | − |
| CAS | − | p = 0.132 | − | − | p = 1.000 | − |
| 4-Chloro-α-methyl | 3.2 | 1.6 | 1 | 12.8 | 12.8 | 2 |
| OG | 0.1 | 0.05 | 0.4 | 0.4 | ||
| 4-Methyl | 12.8 | 6.4 | 1 | 12.8 | 12.8 | 2 |
| OG | 0.1 | 0.05 | 0.4 | 0.4 | ||
| 4-Methoxy | 12.8 | 12.8 | 2 | 12.8 | 12.8 | 2 |
| OG | 0.1 | 0.1 | 0.4 | 0.4 | ||
| Mean: Chemosensitizers | 9.6 | 6.9 | − | 12.8 | 12.8 | − |
| OG | 0.1 | 0.07 | − | 0.4 | 0.4 | − |
| t-test 1: Chemosensitizers | − | p = 0.590 | − | − | p = 1.000 | − |
| OG | − | p = 0.116 | − | − | p = 1.000 | − |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.H.; Chan, K.L.; Cheng, L.W. Cinnamic Acid Analogs as Intervention Catalysts for Overcoming Antifungal Tolerance. Molecules 2017, 22, 1783. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules22101783
Kim JH, Chan KL, Cheng LW. Cinnamic Acid Analogs as Intervention Catalysts for Overcoming Antifungal Tolerance. Molecules. 2017; 22(10):1783. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules22101783
Chicago/Turabian StyleKim, Jong H., Kathleen L. Chan, and Luisa W. Cheng. 2017. "Cinnamic Acid Analogs as Intervention Catalysts for Overcoming Antifungal Tolerance" Molecules 22, no. 10: 1783. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules22101783