Chemoinformatic Database Building and in Silico Hit-Identification of Potential Multi-Targeting Bioactive Compounds Extracted from Mushroom Species
Abstract
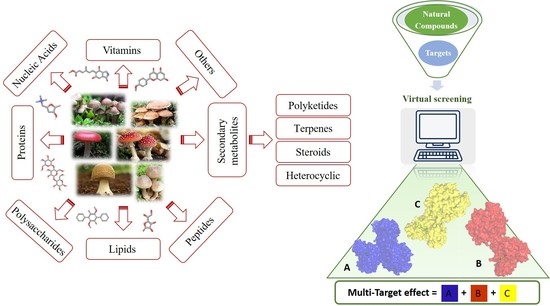
:1. Introduction
2. Results and Discussion
2.1. Multi-Targeted Anticancer Compounds
2.2. Multi-Targeted Anti-Inflammatory Compounds
2.3. Multi-Targeted Compounds in Neurodegenerative Disease
2.4. Multi-Targeted Compounds in Metabolic Diseases
2.5. Non Selective MTAs
3. Materials and Methods
3.1. Database Building
3.2. Database Preparation
3.3. Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries
3.4. Targets Preparation and Clustering
3.5. Docking Simulation
3.6. Data Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chang, S.T.; Miles, P.G. Mushrooms biology—A new discipline. Mycologist 1992, 6, 64–65. [Google Scholar] [CrossRef]
- Hawksworth, D.L. Mushrooms: The extent of the unexplored potential. Int. J. Med. Mushrooms 2001, 3, 333–337. [Google Scholar] [CrossRef]
- Zhang, J.J.; Li, Y.; Zhou, T.; Xu, D.P.; Zhang, P.; Li, S.; Li, H.B. Bioactivities and health benefits of mushrooms mainly from china. Molecules 2016, 21, 938. [Google Scholar] [CrossRef] [PubMed]
- Breene, W. Nutritional and medicinal value of speciality mushrooms. J. Food Prod. Mark. 1990, 53, 883–894. [Google Scholar] [CrossRef]
- Zhong, J.J.; Xiao, J.H. Secondary metabolites from higher fungi: Discovery, bioactivity, and bioproduction. Adv. Biochem. Eng. Biotechnol. 2009, 113, 79–150. [Google Scholar] [CrossRef] [PubMed]
- Leeson, P. The Lipinski rule of five. Nature 2012, 481, 455–456. [Google Scholar] [CrossRef] [PubMed]
- Wasser, S.P. Medicinal mushrooms as a source of antitumor and immunomodulating polysaccharides. Appl. Microbiol. Biotechnol. 2002, 60, 258–274. [Google Scholar] [PubMed]
- Mizuno, T. The extraction and development of antitumour-active polysaccharides from medicinal mushrooms in Japan. Int. J. Med. Mushrooms 1999, 1, 9–29. [Google Scholar] [CrossRef]
- Wang, J.C.; Hu, S.H.; Su, C.H.; Lee, T.M. Antitumor and immunoenhancing activities of polysaccharide from culture broth of Hericium spp. Kaohsiung J. Med. Sci. 2001, 17, 461–467. [Google Scholar] [PubMed]
- Wu, H.T.; Lu, F.H.; Su, Y.C.; Ou, H.Y.; Hung, H.C.; Wu, J.S.; Yang, Y.C.; Chang, C.J. In vivo and in vitro anti-tumor effects of fungal extracts. Molecules 2014, 19, 2546–2556. [Google Scholar] [CrossRef] [PubMed]
- Mau, J.L.; Lin, H.C.; Chen, C.C. Antioxidant properties of several medicinal mushrooms. J. Agric. Food Chem. 2002, 50, 6072–6077. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.Y.; Cho, K.J.; Chung, S.H. Activation of macrophages by GLB, a protein-polysaccharide of the growing tips of Ganoderma lucidum. Yakhak. Hoeji. 1998, 42, 302–306. [Google Scholar]
- Soares, A.A.; de Sá-Nakanishi, A.B.; Bracht, A.; da Costa, S.M.; Koehnlein, E.A.; de Souza, C.G.; Peralta, R.M. Hepatoprotective effects of mushrooms. Molecules 2013, 18, 7609–7630. [Google Scholar] [CrossRef] [PubMed]
- Debnath, T.; Park, S.R.; Kim, D.H.; Jo, J.E.; Lim, B.O.; Debnath, T. Anti-oxidant and anti-inflammatory activities of Inonotus obliquus and germinated brown rice extracts. Molecules 2013, 18, 9293–9304. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, Y.M.; Cao, W.; Yao, K.W.; Liu, Z.Q.; Guo, J.Y. Anti-inflammation and antioxidant effect of cordymin, a peptide purified from the medicinal mushroom Cordyceps sinensis, in middle cerebral artery occlusion-induced focal cerebral ischemia in rats. Metab. Brain Dis. 2012, 27, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Lin, Y.; Luo, Y.L.; Liang, H.H.; Sun, P.L. Extraction, antimicrobial, and antioxidant activities of crude polysaccharides from the wood ear medicinal mushroom Auricularia. auricula-judae (Higher Basidiomycetes). Int. J. Med. Mushrooms 2015, 17, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Bender, S.; Dumitrache, C.N.; Backhaus, J.; Christie, G.; Cross, R.F.; Lonergan, G.T. A case for caution in assessing the antibiotic activity of extracts of culinary-medicinal Shiitake mushroom [Lentinus edodes (Berk.) Singer] (Agaricomycetidae). Int. J. Med. Mushrooms 2003, 5, 31–35. [Google Scholar] [CrossRef]
- Chihara, G.; Chihara, G.; Hamuro, J.; Maeda, Y.Y.; Arai, Y.; Fukuoka, F. Fractionation and purification of the polysaccharides with marked antitumour activity especially leninan from Lentinun edodes. Cancer Res. 1970, 30, 2776–2781. [Google Scholar] [PubMed]
- Wasser, S.P.; Weis, A.L. Medicinal properties of substances occurring in higher basidiomycete mushrooms: Current perspectives. Int. J. Med. Mushrooms 1999, 1, 31–62. [Google Scholar] [CrossRef]
- Taguchi, T.; Furue, H.; Kimura, T.; Kondo, T.; Hattori, T.; Itoh, T.; Ogawa, N. End point result of a randomized controlled study of the treatment of gastrointestinal cancer with a combination of lentinan and chemotherapeutic agents. Excerpta Medica 1985, 40, 151–165. [Google Scholar]
- Kurashiga, S.; Akuzawa, Y.; Eudo, F. Effects of Lentinus edodes, Grifola frondosa and Pleurotus ostreatus administration on cancer outbreaks and activities of macrophages and lymphocytes in mice treated with a carcinogen. Immunopharmacol. Immunotoxicol. 1997, 19, 75–85. [Google Scholar] [CrossRef]
- Chihara, G. Recent progress in immunopharmacology and therapeutic effects of polysaccharides. Dev. Biol. Stand. 1992, 77, 191–197. [Google Scholar] [PubMed]
- Suga, T.; Maeda, Y.Y.; Uchida, H.; Rokutanda, M.; Chihara, G. Macrophage-mediated acute-phase transport protein production induced by Lentinan. Int. J. Immunopharmacol. 1986, 8, 691–699. [Google Scholar] [CrossRef]
- Fruehauf, J.P.; Bonnard, G.D.; Herberman, R.B. The effect of lentinan on production of interleukin-1 by human monocytes. Immunopharmacology. 1982, 5, 65–74. [Google Scholar] [CrossRef]
- Maeda, Y.Y.; Sakaizumi, M.; Moriwaki, K.; Yonekawa, H. Genetic control of the expression of two biological activities of an antitumor polysaccharide, Lentinan. Int. J. Immunopharmacol. 1991, 13, 977–986. [Google Scholar] [CrossRef]
- Miguel, J.; Garcia, B.; Espinosa, M.E.; Ogura, T. Volatile compounds secreted by the oyster mushroom (Pleurotus ostreatus) and their antibacterial activities. J. Agric. Food Chem. 1997, 45, 4049–4052. [Google Scholar]
- Tasaka, K.; Mio, M.; Izushi, K.; Akagi, M.; Makino, T. Anti-allergic constituents in the culture medium of Ganoderma lucidum (II): The inhibitory effect of cyclooctasulfur on histamine release. Agents Actions 1988, 23, 157–160. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.B. Focus on anti-oxidative and free radical scavening activity of Ganoderma lucidum. J. Appl. Pharmacol. 2004, 12, 133–137. [Google Scholar]
- Akihisa, T.; Franzblau, S.G.; Tokuda, H.; Tagata, M. Antitubercular activity and inhibitory effect on Epstein-Barr virus activation of sterols and polyisoprenepolyols from an edible mushroom, Hypsizigus marmoreus. Biol. Pharm. Bull. 2005, 28, 1117–1119. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.K.; Ng, T.B. Hypsin, A novel thermostable ribosome inactivating protein with antifungal and antiproliferative activities from fruiting bodies of the edible mushroom Hypsizigus marmoreus. Biochem. Biophys. Res. Commun. 2001, 285, 1071–1075. [Google Scholar] [CrossRef] [PubMed]
- Konno, S.; Aynehchi, S.; Dolin, D.J.; Schwartz, A.M.; Choudhury, M.S.; Tazakin, H.N. Anticancer and hypoglycemic effects of polysaccharides in edible and medicinal Maitake mushroom [Grifola frondosa (Dicks.:Fr.) S.F.Gray]. Int. J. Med. Mushrooms 2002, 4, 185–195. [Google Scholar] [CrossRef]
- Phan, C.W.; David, P.; Naidu, M.; Wong, K.H.; Sabaratnam, V. Therapeutic potential of culinary-medicinal mushrooms for the management of neurodegenerative diseases: Diversity, metabolite, and mechanism. Crit. Rev. Biotechnol. 2014, 35, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Ganeshpurkar, A.S.; Rai, G.; Jain, A.P. Medicinal mushrooms: Towards a new horizon. Pharmacogn. Rev. 2010, 4, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Jens-Uwe, P. Polypharmacology in Drug Discovery; Hoboken, N.J., Ed.; John Wiley & Sons: Basel, Switzerland, 2012; pp. 9–20. [Google Scholar]
- The Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB). Available online: http://www.rcsb.org/pdb (accessed on 4 August 2017).
- Teiten, M.H.; Eifes, S.; Dicato, M.; Diederich, M. Curcumin-the paradigm of multi-target natural compounds with applications in cancer prevention and treatment. Toxins 2010, 2, 128–162. [Google Scholar] [CrossRef] [PubMed]
- Koeberle, A.; Werz, O. Multi-target approach for natural products in inflammation. Drug. Discov. Today 2014, 19, 1871–1882. [Google Scholar] [CrossRef] [PubMed]
- Engler, M.; Anke, T.; Sterner, O. Pterulinic acid and pterulone, two novel inhibitors of NADH: Ubiquinone oxidoreductase (Complex I) produced by a pterula species. J. Antibiot. 1997, 50, 330–333. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, A.; Bolognesi, M.L.; Minarini, A.; Rosini, M.; Tumiatti, V.; Recanatini, M.; Melchiorre, C. Multi-target-directed ligands to combat neurodegenerative diseases. J. Med. Chem. 2008, 51, 347–372. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Y. One-compound-multiple-targets strategy to combat Alzheimer’s disease. FEBS Lett. 2005, 579, 5260–5264. [Google Scholar] [CrossRef] [PubMed]
- Rosini, M.; Andrisano, V.; Bartolini, M.; Bolognesi, M.L.; Hrelia, P.; Minarini, A.; Tarozzi, A.; Melchiorre, C. Rational approach to discover multipotent anti-Alzheimer drugs. J. Med. Chem. 2005, 48, 360–363. [Google Scholar] [CrossRef] [PubMed]
- Melchiorre, C.; Andrisano, V.; Bolognesi, M.L.; Budriesi, R.; Cavalli, A.; Cavrini, V.; Rosini, M.; Tumiatti, V.; Recanatini, M. Acetylcholinesterase noncovalent inhibitors based on a polyamine backbone for potential use against Alzheimer’s disease. J. Med. Chem. 1998, 41, 4186–4189. [Google Scholar] [CrossRef] [PubMed]
- Ono, K.; Hasegawa, K.; Naiki, H.; Yamada, M. Curcumin has potent anti-amyloidogenic effects for Alzheimer’s β-amyloid fibrils in vitro. Neurosci. Res. 2004, 75, 742–750. [Google Scholar] [CrossRef] [PubMed]
- Gaede, P.; Lund-Andersen, H.; Parving, H.H.; Pedersen, O. Effect of a multifactorial intervention on mortality in type 2 diabetes. N. Engl. J. Med. 2008, 358, 580–591. [Google Scholar] [CrossRef] [PubMed]
- Sheng, H.; Sun, H. Synthesis, biology and clinical significance of pentacyclic triterpenes: A multi-target approach to prevention and treatment of metabolic and vascular diseases. Nat. Prod. Rep. 2011, 28, 543–593. [Google Scholar] [CrossRef] [PubMed]
- Froufe, H.J.C.; Abreu, R.M.V.; Ferreira, I.C.F.R. QCAR models to predict wild mushrooms radical scavenging activity, reducing power and lipid peroxidation inhibition. Chemometr. Intell. Lab. 2011, 109, 192–196. [Google Scholar] [CrossRef]
- Bawa, P.; Pradeep, P.; Kumar, P.; Choonara, Y.E.; Modi, G.; Pillay, V. Multi-target therapeutics for neuropsychiatric and neurodegenerative disorders. Drug Discov. Today 2016, 21, 1886–1914. [Google Scholar] [CrossRef] [PubMed]
- Oshida, M.; Matsuura, Y.; Hotta, S.; Watanabe, J.; Mogi, Y.; Watanabe, T. Isolation and identification of a humanTRPV1 activating compound from soy sauce. Biosci. Biotechnol. Biochem. 2017, 81, 987–994. [Google Scholar] [CrossRef] [PubMed]
- National Centre for Biotechnological Information (NCBI). Available online: https://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/ (accessed on 4 August 2017).
- PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/ (accessed on 4 August 2017).
- ChEMBL. Available online: https://www.ebi.ac.uk/chembl/ (accessed on 4 August 2017).
- ChemSpider. Available online: http://www.chemspider.com/ (accessed on 4 August 2017).
- MarvinSketch. Available online: https://www.chemaxon.com/products/marvin/marvinsketch/ (accessed on 18 September 2017).
- Instant Jchem. Available online: https://www.chemaxon.com/products/instant-jchem-suite/instant-jchem/ (accessed on 4 August 2017).
- LigPrep; Schrödinger, LLC: New York, NY, USA, 2017.
- Maestro; Schrödinger, LLC: New York, NY, USA, 2017.
- Schrödinger Suites; Schrödinger, LLC: New York, NY, USA, 2017.
- Jonathan, B.; Michael, A.W. Chemical con artists foil drug discovery. Nature 2014, 513, 481–483. [Google Scholar] [CrossRef]
- ZINC15. Available online: http://zinc15.docking.org/patterns/home (accessed on 4 August 2017).
- CBLIGAND. Available online: http://www.cbligand.org/PAINS/ (accessed on 4 August 2017).
- FAFDRUGS3. Available online: http://fafdrugs3.mti.univ-paris-diderot.fr/ (accessed on 4 August 2017).
- ADVISOR. Available online: http://advisor.docking.org (accessed on 4 August 2017).
- Aldrich, C.; Bertozzi, C.; Georg, G.I.; Kiessling, L.; Lindsley, C.; Liotta, D.; Merz, K.M.; Schepartz, A.; Wang, S. The ecstasy and agony of assay interference compounds. ACS Med. Chem. Lett. 2017, 4, 379–382. [Google Scholar] [CrossRef] [PubMed]
- Protein Preparation Wizard; Schrödinger, LLC: New York, NY, USA, 2017.
- Glide; Schrödinger, LLC: New York, NY, USA, 2017.
- Medina-Franco, J.L.; Giulianotti, M.A.; Welmaker, G.S.; Houghten, R.A. Shifting from the single to the multitarget paradigm in drug discovery. Drug Discov. Today 2013, 18, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Chemotheca. Available online: http://www.unicz.chemoteca.it (accessed on 8 August 2017).
- COST Action CA15135. Available online: http://www.mutalig.eu/ (accessed on 8 August 2017).
Sample Availability: Not Available. |












| Name | Structure | Targets | G-Score (kcal/mol) |
|---|---|---|---|
| Inotopyrrole |  | c-Met | −8.05 |
| VEGFR2 | −8.08 | ||
| EGFR | −8.22 | ||
| MEK2 | −8.09 | ||
| Marasmone 21 |  | c-Met | −8.34 |
| MEK1 | −8.02 | ||
| Fascicularone A |  | c-Met | −8.51 |
| SIRT1 | −8.78 | ||
| Enokipodin H |  | c-Met | −8.79 |
| GSK3β | −8.21 | ||
| SIRT1 | −8.40 | ||
| Confluentin |  | VEGFR2 | −10.16 |
| EGFR | −10.16 | ||
| PDK1 | −8.97 | ||
| SIRT1 | −8.75 | ||
| Blennin C |  | c-Met | −10.16 |
| MEK1 | −8.97 | ||
| SIRT1 | −8.75 | ||
| Acromelic acid A |  | PDK1 | −8.50 |
| CA IX | −8.22 | ||
| CA XII | −8.32 | ||
| Ganomycin B |  | VEGFR2 | −8.44 |
| B-RAF V600E | −8.29 | ||
| PDK1 | −8.74 | ||
| SIRT1 | −8.19 | ||
| Atromentic acid |  | c-Met | −8.51 |
| VEGFR2 | −9.09 | ||
| B-RAF wt | −8.5 | ||
| PDK1 | −8.14 | ||
| 1-O-Acetyl-3-epi-illudol |  | c-Met | −8.97 |
| MEK1 | −9.37 | ||
| PDK1 | −8.25 | ||
| GSK3β | −8.4 | ||
| Gentianal |  | c-Met | −8.77 |
| VEGFR2 | −8.5 | ||
| EGFR | −8.27 | ||
| B-RAF V600E | −8.08 | ||
| BMX | −8.02 | ||
| Hericenone H |  | c-Met | −8.82 |
| EGFR | −8.48 | ||
| Hericenol A |  | c-Met | −8.68 |
| MEK1 | −8.00 | ||
| MEK2 | −8.17 | ||
| SIRT1 | −8.46 | ||
| Erinacerin P |  | MEK1 | −8.48 |
| SGK1 | −8.06 | ||
| Erinacerin A |  | EGFR | −8.2 |
| B-RAF wt | −8.48 | ||
| SIRT1 | −8.36 | ||
| Hericenol B |  | c-Met | −9.52 |
| EGFR | −8.41 | ||
| MEK1 | −8.66 | ||
| MEK2 | −8.89 | ||
| SIRT1 | −9.11 | ||
| Genistin |  | VEGFR2 | −8.22 |
| B-RAF wt | −8.91 | ||
| MEK1 | −9.52 | ||
| SGK1 | −8.39 | ||
| Illudinine |  | Aurora A kinase | −8.59 |
| c-Met | −8.89 | ||
| VEGFR2 | −8.79 | ||
| B-RAF V600E | −8.9 | ||
| BMX | −8.13 | ||
| GSK3β | −8.67 | ||
| SIRT1 | −8.11 | ||
| Daldinal |  | c-Met | −8.62 |
| VEGFR2 | −8.85 | ||
| B-RAF V600E | −8.03 | ||
| PI3Kγ | −8.34 | ||
| Narigin |  | ERK1 | −8.42 |
| B-RAF wt | −9.89 | ||
| B-RAF V600E | −9.64 | ||
| MEK1 | −9.12 | ||
| BMX | −10.04 | ||
| SGK1 | −8.99 | ||
| (R)-Torosachrysone |  | c-Met | −8.96 |
| VEGFR2 | −8.69 | ||
| ERK1 | −8.29 | ||
| MEK1 | −8.53 | ||
| PI3Kα | −8.06 | ||
| BMX | −8.22 | ||
| GSK3β | −8.55 | ||
| SIRT1 | −8.04 |
| Name | Structure | Target | G-Score (kcal/mol) |
|---|---|---|---|
| Illudacetalic Acid |  | COX-2 | −8.42 |
| Adenosine A2A R | −8.09 | ||
| Pterulinic Acid |  | COX-1 | −8.97 |
| Glucocorticoid R | −8.73 | ||
| Orellanine |  | COX-1 | −8.64 |
| COX-2 | −8.85 | ||
| Glucocorticoid R | −8.39 | ||
| Bisnoryangonin |  | COX-2 | −8.17 |
| Glucocorticoid R | −8.34 | ||
| 1-Naphthylacetic acid |  | COX-1 | −9.33 |
| COX-2 | −8.33 | ||
| Pulvinic acid |  | COX-2 | −8.56 |
| Glucocorticoid R | −8.23 |
| Name | Structure | Targets | G-Score (kcal/mol) |
|---|---|---|---|
| Erinacerin O |  | AChE | −8.70 |
| COMT | −8.00 | ||
| Cordysinin A |  | GSK3β | −8.13 |
| AChE | −8.47 | ||
| COX-2 | −8.34 | ||
| Hesperidin |  | AChE | −10.67 |
| COMT | −9.18 | ||
| AdenosineA2A R | −9.63 | ||
| Vitexin |  | AChE | −8.93 |
| COMT | −8.09 | ||
| Pterulone |  | AChE | −8.38 |
| MAO-B | −8.16 | ||
| COX-2 | −8.54 | ||
| GSK3β | −8.09 |
| Name | Structure | Targets | G-Score (kcal/mol) |
|---|---|---|---|
| Hericenone A |  | Insulin R | −8.62 |
| IGFR | −8.23 | ||
| PPAR-α | −8.49 | ||
| PPAR-γ | −8.81 | ||
| GSK3β | −8.01 | ||
| O-xylotocopherol |  | PPAR-α | −9.00 |
| PPAR-γ | −9.20 | ||
| 3,4-Dihydro-2-methyl-2-(2-oxo-4-methyl-3-pentenyl)-5-methoxy-2H-furo[3,4-h][1]benzopyran-9(7H)-one |  | PPAR-α | −8.05 |
| PPAR-γ | −8.27 | ||
| 7-Acetyl-4-methylazulene-1-carboxylic acid |  | Insulin R | −8.12 |
| PPAR-α | −8.43 | ||
| 4,6-Dihydroxyisobenzofuran-1,3-dione |  | PKA C-α | −8.84 |
| PPAR-γ | −8.49 | ||
| GSK3β | −8.10 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maruca, A.; Moraca, F.; Rocca, R.; Molisani, F.; Alcaro, F.; Gidaro, M.C.; Alcaro, S.; Costa, G.; Ortuso, F. Chemoinformatic Database Building and in Silico Hit-Identification of Potential Multi-Targeting Bioactive Compounds Extracted from Mushroom Species. Molecules 2017, 22, 1571. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules22091571
Maruca A, Moraca F, Rocca R, Molisani F, Alcaro F, Gidaro MC, Alcaro S, Costa G, Ortuso F. Chemoinformatic Database Building and in Silico Hit-Identification of Potential Multi-Targeting Bioactive Compounds Extracted from Mushroom Species. Molecules. 2017; 22(9):1571. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules22091571
Chicago/Turabian StyleMaruca, Annalisa, Federica Moraca, Roberta Rocca, Fulvia Molisani, Francesca Alcaro, Maria Concetta Gidaro, Stefano Alcaro, Giosuè Costa, and Francesco Ortuso. 2017. "Chemoinformatic Database Building and in Silico Hit-Identification of Potential Multi-Targeting Bioactive Compounds Extracted from Mushroom Species" Molecules 22, no. 9: 1571. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules22091571