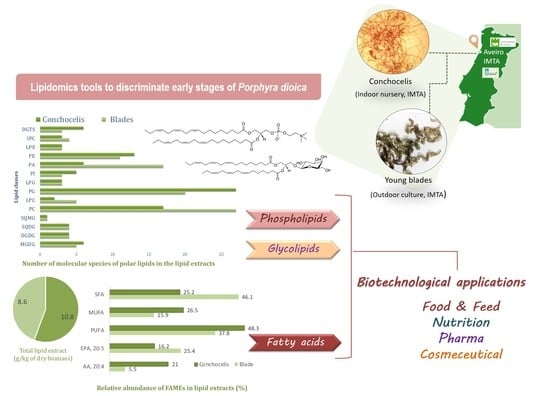
High-Resolution Lipidomics of the Early Life Stages of the Red Seaweed Porphyra dioica
Abstract
:1. Introduction
2. Results and Discussion
2.1. Fatty Acids Profile from Porphyra Dioica
2.2. Polar Lipids from Porphyra dioica
2.2.1. Glycolipids—GLs
2.2.2. Phospholipids (PLs)
2.2.3. Betaine Lipids
3. Experimental Section
3.1. Materials
3.2. Biomass
3.3. Total Lipid Extraction
3.4. Analysis of Fatty Acid Methyl Esters—FAMEs by Gas Chromatography–Mass Spectrometry (GC–MS)
3.5. Analysis of Polar Lipids by Hydrophilic Interaction Liquid Chromatography–Mass Spectrometry Based (LC–MS)
3.6. Nutritional Values
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sahoo, S.; Tang, X.; Yarish, C. Porphyra—The economic seaweed as a new experimental system. Curr. Sci. 2002, 83, 1313–1316. [Google Scholar]
- Blouin, N.A.; Brodie, J.A.; Grossman, A.C.; Xu, P.; Brawley, S.H. Porphyra: A marine crop shaped by stress. Trends Plant Sci. 2011, 16, 29–37. [Google Scholar] [CrossRef] [PubMed]
- WoRMS Editorial Board. World Register of Marine Species. 2017. Available online: http://www.marinespecies.org at VLIZ (accessed on 20 January 2017). [CrossRef]
- Pereira, R.; Kraemer, G.; Yarish, C.; Sousa-Pinto, I. Nitrogen uptae by gametophytes of Porphyra dioica (Bangiales, Rhodophyta) under controlled-culture conditions. Eur. J. Phycol. 2008, 43, 107–118. [Google Scholar] [CrossRef]
- Pereira, R.; Sousa-Pinto, I.; Yarish, C. Field and culture studies of the life history of Porphyra dioica (Bangiales, Rhodophyta) from Portugal. Phycologia 2004, 43, 756–767. [Google Scholar] [CrossRef]
- Drew, K.M. Studies in the Bangioideae. Ann. Bot. 1954, 18, 183–216. [Google Scholar] [CrossRef]
- Candia, A.; Lindstrom, S.; Reyes, E. Porphyra sp. (Bangiales, Rhodophyta): Reproduction and life form. Hydrobiologia 1999, 398, 115–119. [Google Scholar] [CrossRef]
- Stengel, D.B.; Connan, S.; Popper, Z.A. Algal chemodiversity and bioactivity: Sources of natural variability and implications for commercial application. Biotechnol. Adv. 2011, 29, 483–501. [Google Scholar] [CrossRef] [PubMed]
- Stabili, L.; Acquaviva, M.I.; Biandolino, F.; Cavallo, R.A.; De Pascali, S.A.; Fanizzi, F.P.; Narracci, M.; Petrocelli, A.; Cecere, E. The lipidic extract of the seaweed Gracilariopsis longissima (Rhodophyta, Gracilariales): A potential resource for biotechnological purposes? New Biotechnol. 2012, 29, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Gao, J.T.; Zhang, Y.C.; Qin, S. Optimization of conditions for cell cultivation of Porphyra haitanensis conchocelis in a bubble-column bioreactor. World J. Microbiol. Biotechnol. 2006, 22, 655–660. [Google Scholar] [CrossRef]
- Van Ginneken, V.J.T.; Helsper, J.P.F.G.; de Visser, W.; Van Keulen, H.; Brandenburg, W.A. Polyunsaturated fatty acids in various macroalgal species from North Atlantic and tropical seas. Lipids Health Dis. 2011, 10, 104. [Google Scholar] [CrossRef] [PubMed]
- Harwood, J.L.; Guschina, I.A. The versatility of algae and their lipid metabolism. Biochimie 2009, 91, 679–684. [Google Scholar] [CrossRef] [PubMed]
- Plouguerné, E.; da Gama, B.A.P.; Pereira, R.C.; Barreto-Bergter, E. Glycolipids from seaweeds and their potential biotechnological applications. Front. Cell. Infect. Microbiol. 2014, 4, 174. [Google Scholar] [CrossRef] [PubMed]
- Banskota, A.H.A.H.; Stefanova, R.; Sperker, S.; Lall, S.; Craigie, J.S.J.S.; Hafting, J.T.J.T. Lipids isolated from the cultivated red alga Chondrus crispus inhibit nitric oxide production. J. Appl. Phycol. 2014, 26, 1565–1571. [Google Scholar] [CrossRef]
- Banskota, A.H.; Stefanova, R.; Sperker, S.; Lall, S.P.; Craigie, J.S.; Hafting, J.T.; Critchley, A.T. Polar lipids from the marine macroalga Palmaria palmata inhibit lipopolysaccharide-induced nitric oxide production in RAW264.7 macrophage cells. Phytochemistry 2014, 101, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Robertson, R.C.; Guihéneuf, F.; Bahar, B.; Schmid, M.; Stengel, D.B.; Fitzgerald, G.F.; Ross, R.P.; Stanton, C. The anti-Inflammatory effect of algae-derived lipid extracts on lipopolysaccharide (LPS)-stimulated human THP-1 macrophages. Mar. Drugs 2015, 13, 5402–5424. [Google Scholar] [CrossRef] [PubMed]
- Eitsuka, T.; Nakagawa, K.; Igarashi, M.; Miyazawa, T. Telomerase inhibition by sulfoquinovosyldiacylglycerol from edible purple laver (Porphyra yezoensis). Cancer Lett. 2004, 212, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Chou, H.N. Screening of red algae filaments as a potential alternative source of eicosapentaenoic acid. Mar. Biotechnol. 2002, 4, 189–192. [Google Scholar] [CrossRef] [PubMed]
- Xiaolei, F.; Guangce, W.; Demao, L.; Pu, X.; Songdong, S. Study on early-stage development of conchospore in Porphyra yezoensis Ueda. Aquaculture 2008, 278, 143–149. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, P.; Luo, Q.; Yan, X.; Xu, J.; Chen, J.; Chen, H. Metabolite changes during the life history of Porphyra haitanensis. Plant Biol. 2015, 17, 660–666. [Google Scholar] [CrossRef] [PubMed]
- Spolaore, P.; Joannis-Cassan, C.; Duran, E.; Isambert, A. Commercial applications of microalgae. J. Biosci. Bioeng. 2006, 101, 87–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soler-Vila, A.; Coughlan, S.; Guiry, M.D.; Kraan, S. The red alga Porphyra dioica as a fish-feed ingredient for rainbow trout (Oncorhynchus mykiss): Effects on growth, feed efficiency, and carcass composition. J. Appl. Phycol. 2009, 21, 617–624. [Google Scholar] [CrossRef]
- Rulong, L. Pyropia conchocelis: Potential as an algal source for carotenoid extraction. Am. J. BioSci. 2015, 3, 121–132. [Google Scholar] [CrossRef]
- Leal, M.C.; Munro, M.H.G.; Blunt, J.W.; Puga, J.; Jesus, B.; Calado, R.; Rosa, R.; Madeira, C. Biogeography and biodiscovery hotspots of macroalgal marine natural products. Nat. Prod. Rep. 2013, 30, 1380–1390. [Google Scholar] [CrossRef] [PubMed]
- Bhakuni, D.S.; Rawat, D.S. Bioactive metabolites of marine algae, fungi and bacteria. In Bioactive Marine Natural Products; Springer: Berlin, Germany, 2005; pp. 1–19. ISBN 1402034725. [Google Scholar]
- Wu, D.; Fujio, M.; Wong, C.H. Glycolipids as immunostimulating agents. Bioorgan. Med. Chem. 2008, 16, 1073–1083. [Google Scholar] [CrossRef] [PubMed]
- Burri, L.; Hoem, N.; Banni, S.; Berge, K. Marine omega-3 phospholipids: Metabolism and biological activities. Int. J. Mol. Sci. 2012, 13, 15401–15419. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C. Marine omega-3 fatty acids and inflammatory processes: Effects, mechanisms and clinical relevance. Biochim. Biophys. Acta 2014, 1851, 469–484. [Google Scholar] [CrossRef] [PubMed]
- El Baky, H.H.A.; El Baz, F.K.; El Baroty, G.S.; Asker, M.M.S.; Ibrahim, E.A. Phospholipids of some marine microalgae: Identification, antivirus, anticancer and antimicrobial bioactivities. Der Pharma Chem. 2014, 6, 9–18. [Google Scholar]
- Lu, F.S.H.; Nielsen, N.S.; Baron, C.P.; Jacobsen, C. Marine phospholipids: The current understanding of their oxidation mechanisms and potential uses for food fortification. Crit. Rev. Food Sci. Nutr. 2017, 57, 2057–2070. [Google Scholar] [CrossRef] [PubMed]
- Cian, R.E.; Fajardo, M.A.; Alaiz, M.; Vioque, J.; González, R.J.; Drago, S.R. Chemical composition, nutritional and antioxidant properties of the red edible seaweed Porphyra columbina. Int. J. Food Sci. Nutr. 2014, 65, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Machado, D.I.; López-Cervantes, J.; López-Hernández, J.; Paseiro-Losada, P. Fatty acids, total lipid, protein and ash contents of processed edible seaweeds. Food Chem. 2004, 85, 439–444. [Google Scholar] [CrossRef]
- Schmid, M.; Guihéneuf, F.; Stengel, D.B. Fatty acid contents and profiles of 16 macroalgae collected from the Irish Coast at two seasons. J. Appl. Phycol. 2014, 26, 451–463. [Google Scholar] [CrossRef]
- Fleurence, J.; Gutbier, G.; Mabeau, S.; Leray, C. Fatty acids from 11 marine macroalgae of the French Brittany coast. J. Appl. Phycol. 1994, 6, 527–532. [Google Scholar] [CrossRef]
- Luo, Q.; Zhu, Z.; Zhu, Z.; Yang, R.; Qian, F.; Chen, H.; Yan, X. Different responses to heat shock stress revealed heteromorphic adaptation strategy of Pyropia haitanensis (Bangiales, Rhodophyta). PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Simopoulos, A. The importance of the ratio of omega-6/omega-3 essential fatty acids. Biomed. Pharmacother. 2002, 56, 365–379. [Google Scholar] [CrossRef]
- Kumari, P.; Reddy, C.R.K.; Jha, B. Comparative evaluation and selection of a method for lipid and fatty acid extraction from macroalgae. Anal. Biochem. 2011, 415, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C.; Albers, R.; Antoine, J.-M.; Blum, S.; Ferns, G.A.; Folkerts, G.; Bourdet-Sicard, R.; Friedmann, P.S.; Frost, G.S.; Guarner, F.; et al. Inflammatory disease processes and interactions with nutrition. Br. J. Nutr. 2009, 101, S2–S45. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C. Omega-3 polyunsaturated fatty acids and inflammatory processes: Nutrition or pharmacology? Br. J. Clin. Pharmacol. 2013, 75, 645–662. [Google Scholar] [CrossRef] [PubMed]
- De Roos, B.; Mavrommatis, Y.; Brouwer, I.A. Long-chain n-3 polyunsaturated fatty acids: New insights into mechanisms relating to inflammation and coronary heart disease. Br. J. Pharmacol. 2009, 158, 413–428. [Google Scholar] [CrossRef] [PubMed]
- Lordan, S.; Ross, R.P.; Stanton, C. Marine bioactives as functional food ingredients: Potential to reduce the incidence of chronic diseases. Mar. Drugs 2011, 9, 1056–1100. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C. Omega-3 fatty acids and inflammatory processes: From molecules to man. Biochem. Soc. Trans. 2017, 1105–1115. [Google Scholar] [CrossRef] [PubMed]
- Pereira, H.; Barreira, L.; Figueiredo, F.; Custódio, L.; Vizetto-Duarte, C.; Polo, C.; Rešek, E.; Engelen, A.; Varela, J. Polyunsaturated Fatty acids of marine macroalgae: Potential for nutritional and pharmaceutical applications. Mar. Drugs 2012, 10, 1920–1935. [Google Scholar] [CrossRef] [PubMed]
- Simopoulos, A.P. The importance of the omega-6/omega-3 fatty acid ratio in cardiovascular disease and other chronic diseases. Exp. Biol. Med. 2008, 233, 674–688. [Google Scholar] [CrossRef] [PubMed]
- Simopoulos, A.P. Essential fatty acids in health and chronic disease. Am. J. Clin. Nutr. 1999, 70, 560S–569S. [Google Scholar] [PubMed]
- Ulbricht, T.L.V.; Southgate, D.A.T. Coronary heart disease: Seven dietary factors. Lancet 1991, 338, 985–992. [Google Scholar] [CrossRef]
- Kumar, M.; Kumari, P.; Trivedi, N.; Shukla, M.K.; Gupta, V.; Reddy, C.R.K.; Jha, B. Minerals, PUFAs and antioxidant properties of some tropical seaweeds from Saurashtra coast of India. J. Appl. Phycol. 2010, 23, 797–810. [Google Scholar] [CrossRef]
- Guschina, I.A.; Harwood, J.L. Lipids and lipid metabolism in eukaryotic algae. Prog. Lipid Res. 2006, 45, 160–186. [Google Scholar] [CrossRef] [PubMed]
- Khotimchenko, S.V. Distribution of glyceroglycolipids in marine algae and grasses. Chem. Nat. Compd. 2002, 38, 186–191. [Google Scholar] [CrossRef]
- Melo, T.; Alves, E.; Azevedo, V.; Martins, A.S.; Neves, B.; Domingues, P.; Calado, R.; Abreu, M.H.; Domingues, M.R. Lipidomics as a new approach for the bioprospecting of marine macroalgae—Unraveling the polar lipid and fatty acid composition of Chondrus crispus. Algal Res. 2015, 8, 181–191. [Google Scholar] [CrossRef]
- Da Costa, E.; Melo, T.; Moreira, A.; Bernardo, C.; Helguero, L.; Ferreira, I.; Cruz, M.; Rego, A.; Domingues, P.; Calado, R.; et al. Valorization of lipids from Gracilaria sp. through lipidomics and decoding of antiproliferative and anti-Inflammatory activity. Mar. Drugs 2017, 15, 62. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, E.; Melo, T.; Moreira, A.S.P.; Alves, E.; Domingues, P.; Calado, R.; Abreu, M.H.M.H.; Domingues, M.R. Decoding bioactive polar lipid profile of the macroalgae Codium tomentosum from a sustainable IMTA system using a lipidomic approach. Algal Res. 2015, 12, 388–397. [Google Scholar] [CrossRef]
- Kumari, P.; Reddy, R.; Jha, B. Quantification of selected endogenous hydroxy-oxylipins from tropical marine macroalgae. Mar. Biotechnol. 2014, 16, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Khozin-Goldberg, I.; Cohen, Z. Unraveling algal lipid metabolism: Recent advances in gene identification. Biochimie 2011, 93, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Benning, C. Chloroplast lipid synthesis and lipid trafficking through ER-plastid membrane contact sites. Biochem. Soc. Trans. 2012, 40, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.; Pan, B.S. Identification of sulfoglycolipid bioactivities and characteristic fatty acids of marine macroalgae. JAFC 2012, 60, 8404–8410. [Google Scholar] [CrossRef] [PubMed]
- Mühlroth, A.; Li, K.; Røkke, G.; Winge, P.; Olsen, Y.; Hohmann-Marriott, M.F.; Vadstein, O.; Bones, A.M. Pathways of lipid metabolism in marine algae, co-expression network, bottlenecks and candidate genes for enhanced production of EPA and DHA in species of Chromista. Mar. Drugs 2013, 11, 4662–4697. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Su, X.; Luo, Q.; Xu, J.; Chen, J.; Yan, X.; Chen, H. Profiles of glycerolipids in Pyropia haitanensis and their changes responding to agaro-oligosaccharides. J. Appl. Phycol. 2014, 2397–2404. [Google Scholar] [CrossRef]
- Birner, R.; Bürgermeister, M.; Schneiter, R.; Daum, G. Roles of phosphatidylethanolamine and of its several biosynthetic pathways in Saccharomyces cerevisiae. Mol. Biol. Cell 2001, 12, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Thompson, G.A. Lipids and membrane function in green algae. Biochim. Biophys. Acta Lipids Lipid Metab. 1996, 1302, 17–45. [Google Scholar] [CrossRef]
- Gerasimenko, N.I.; Busarova, N.G.; Moiseenko, O.P. Age-dependent changes in the content of lipids, fatty acids, and pigments in brown alga Costaria costata. Russ. J. Plant Physiol. 2010, 57, 62–68. [Google Scholar] [CrossRef]
- Munnik, T. Phosphatidic acid: An emerging plant lipid second messenger. Trends Plant Sci. 2001, 6, 227–233. [Google Scholar] [CrossRef]
- Testerink, C.; Munnik, T. Phosphatidic acid: A multifunctional stress signaling lipid in plants. Trends Plant Sci. 2005, 10, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Dembitsky, V.M.; Rozentsvet, O.A. Phospholipid composition of some marine red algae. Phytochemistry 1990, 29, 3149–3152. [Google Scholar] [CrossRef]
- Mueller-Roeber, B. Inositol Phospholipid Metabolism in Arabidopsis. Characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol. 2002, 130, 22–46. [Google Scholar] [CrossRef] [PubMed]
- Munnik, T.; Nielsen, E. Green light for polyphosphoinositide signals in plants. Curr. Opin. Plant Biol. 2011, 14, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Michell, R.H. Inositol and its derivatives: Their evolution and functions. Adv. Enzyme Regul. 2011, 51, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Khotimchenko, S.V. Lipids from the marine alga Gracilaria verrucosa. Chem. Nat. Compd. 2005, 41, 285–288. [Google Scholar] [CrossRef]
- Khotimchenko, S.V. Variations in lipid composition among different developmental stages of Gracilaria verrucosa (Rhodophyta). Bot. Mar. 2006, 49, 34–38. [Google Scholar] [CrossRef]
- Paul-André, S.; Norio, M.; Joyard, J.; Maréchal, E.; Miège, C.; Block, M.; Dorne, A.-J.; Douce, R. Structure, Distribution and biosynthesis of glycerolipids from higher plant chloroplasts. Lipids Photosynth. Struct. Funct. Genet. 2004, 6, 21–52. [Google Scholar] [CrossRef]
- Levchenko, E.V. Carbon metabolism transitions during the development of marine macroalga Gracilaria verrucosa. Russ. J. Plant Physiol. 2003, 50, 68–71. [Google Scholar] [CrossRef]
- Kunzler, K.; Eichenberger, W. Betaine lipids and zwitterionic phospholipids in plants and fungi. Phytochemistry 1997, 46, 883–892. [Google Scholar] [CrossRef]
- Sato, N. Betaine Lipids. Bot. Mag. Tokyo 1992, 1, 185–197. [Google Scholar] [CrossRef]
- Kato, M.; Sakai, M.; Adachi, K.; Ikemoto, H.; Sano, H. Distribution of betaine lipids in marine algae. Phytochemistry 1996, 42, 1341–1345. [Google Scholar] [CrossRef]
- Dembitsky, V.M. Betaine ether-linked glycerolipids: Chemistry and biology. Prog. Lipid Res. 1996, 35, 1–51. [Google Scholar] [CrossRef]
- Li, S.; Xu, J.; Chen, J.J.; Chen, J.J.; Zhou, C.; Yan, X. The major lipid changes of some important diet microalgae during the entire growth phase. Aquaculture 2014, 428–429, 104–110. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rappid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Aued-Pimentel, S.; Lago, J.H.G.; Chaves, M.H.; Kumagai, E.E. Evaluation of a methylation procedure to determine cyclopropenoids fatty acids from Sterculia striata St. Hil. Et Nauds seed oil. J. Chromatogr. A 2004, 1054, 235–239. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the seaweeds and lipid extracts are available from authors (or from MDPI). |





| Blade | Conchocelis | |||
|---|---|---|---|---|
| Fatty Acids | Mean (mg kg−1 Dry Biomass) ± SD | (Mean %) | Mean (mg kg−1 Dry Biomass) ± SD | (Mean %) |
| 14:0 | 12.32 ± 0.00 | (0.60) | 28.84 ± 0.83 | (0.81) |
| 15:0 | 20.04 ± 0.34 | (0.98) | 35.24 ± 0.13 | (0.98) |
| 16:0 | 882.1 ± 12.4 | (43.2) | 784.7 ± 21.1 | (21.9) |
| 18:0 | 27.85 ± 0.26 | (1.36) | 52.26 ± 8.00 | (1.46) |
| Σ SFA | 942.3 | 46.1 | 901.0 | 25.2 |
| 16:1(n-7) | 39.83 ± 0.42 | (1.95) | 210.3 ± 5.06 | (5.88) |
| 18:1(n-9) | 184.8 ± 17.7 | (9.04) | 366.1 ± 50.0 | (10.2) |
| 18:1(n-7) | 44.27 ± 1.33 | (2.17) | 287.2 ± 22.7 | (8.03) |
| 20:1(n-9) | 51.63 ± 1.33 | (2.53) | 76.75 ± 8.76 | (2.14) |
| 22:1(n-9) | 4.972 ± 0.04 | (0.24) | 7.953 ± 1.26 | (0.22) |
| Σ MUFA | 325.5 | 15.9 | 948.3 | 26.5 |
| 18:2(n-6) | 81.89 ± 4.06 | (4.01) | 171.6 ± 18.6 | (4.80) |
| 18:3(n-3) | 16.62 ± 0.05 | (0.46) | ||
| 20:2(n-6) | 18.44 ± 0.98 | (0.90) | 30.56 ± 2.70 | (0.85) |
| 20:3(n-6) | 44.82 ± 2.27 | (2.19) | 179.8 ± 17.8 | (5.02) |
| 20:4(n-6) | 112.4 ± 4.22 | (5.50) | 751.0 ± 87.3 | (21.0) |
| 20:5(n-3) | 518.3 ± 14.2 | (25.4) | 579.5 ± 96.4 | (16.2) |
| Σ PUFA | 775.9 | 38.0 | 1729 | 48.3 |
| Total | 2044 | 3578 | ||
| n-6/n-3 | 0.50 | 1.90 | ||
| IA * | 0.85 | 0.34 | ||
| IT * | 0.49 | 0.30 |
| Galactolipids [M + NH4]+ | Fatty Acyl Composition | ||
| m/z Theoretical | (C:N) | Blade | Conchocelis |
| 772.594 | MGDG (34:2) | 18:2/16:0 | 18:2/16:0 |
| 774.609 | MGDG (34:1) | 18:1/16:0 | 18:1/16:0 |
| 794.578 | MGDG (36:5) | 20:5/16:0 | 20:5/16:0 |
| 796.594 | MGDG (36:4) | 20:4/16:0 | 20:4/16:0 |
| 800.624 | MGDG (36:2) | 18:2/18:0 | 18:2/18:0 |
| 802.641 | MGDG (36:1) | ----------- | 18:1/18:0 |
| 934.647 | DGDG (34:2) | 18:2/16:0 | 18:2/16:0 |
| 936.662 | DGDG (34:1) | 18:1/16:0 | 18:1/16:0 |
| 956.631 | DGDG (36:5) | 20:5/16:0 | 20:5/16:0 |
| 972.626 | DGDG (36:5-OH) | 20:5-OH/16:0 | 20:5-OH/16:0 |
| Sulfolipids [M − H]− | Fatty Acyl Composition | ||
| m/z Theoretical | (C:N) | Blade | Conchocelis |
| 555.284 | SQMG 16:0 | 16:0 | 16:0 |
| 793.514 | SQDG (32:0) | 16:0/16:0 | 16:0/16:0 |
| 817.514 | SQDG (34:2) | 18:2/16:0 | 18:2/16:0 |
| 839.498 | SQDG (36:5) | 20:5/16:0 | 20:5/16:0 |
| 855.493 | SQDG (36:5-OH) | 20:5-OH/16:0 | 20:5-OH/16:0 |
| [M + H]+ | Fatty Acyl Composition | ||
|---|---|---|---|
| m/z Theoretical | (C:N) | Blade | Conchocelis |
| Phosphatidylcholine (PC) | |||
| 732.554 | PC (32:1) | 16:0/16:1 and 14:0/18:1 | ----------- |
| 734.569 | PC (32:0) | 16:0/16:0 and 14:0/18:0 | 16:0/16:0 |
| 752.523 | PC (34:5) | ----------- | 14:0/20:5 |
| 754.538 | PC (34:4) | 14:0/20:4 and 16:2/18:2 | 14:0/20:4 and 16:2/18:2 |
| 756.554 | PC (34:3) | 16:0/18:3 and 14:0/20:3 | 16:1/18:2 |
| 758.569 | PC (34:2) | 16:0/18:2 and 16:1/18:1 | 16:0/18:2 and 16:1/18:1 |
| 760.585 | PC (34:1) | 16:0/18:1 | 16:0/18:1 |
| 762.601 | PC (34:0) | 16:0/18:0 | ----------- |
| 780.554 | PC (36:5) | 16:0/20:5 and 16:1/20:4 | 16:0/20:5 |
| 782.569 | PC (36:4) | 16:0/20:4 | 16:0/20:4 |
| 784.585 | PC (36:3) | 16:0/20:3 and 18:1/18:2 | 16:0/20:3 and 18:1/18:2 |
| 786.601 | PC (36:2) | 18:1/18:1 and 18:0/18:2 | 18:1/18:1 and 18:0/18:2 |
| 788.616 | PC (36:1) | 18:0/18:1 | ----------- |
| 804.554 | PC (38:7) | 18:3/20:4 and 18:2/20:5 | ----------- |
| 806.569 | PC (38:6) | 18:2/20:4 and 18:1/20:5 | 18:1/20:5 |
| 808.585 | PC (38:5) | 18:1/20:4 | 18:1/20:4 |
| 810.601 | PC (38:4) | 18:1/20:3 | 18:1/20:3 |
| 812.616 | PC (38:3) | 18:0/20:3 | ----------- |
| Lyso-phosphatidylcholine (LPC) | |||
| 494.339 | LPC (16:1) | LPC (16:1) | ------------_ |
| 496.323 | LPC (16:0) | LPC (16:0) | LPC (16:0) |
| 518.321 | LPC (18:3) | LPC (18:3) | ------------ |
| 522.355 | LPC (18:1) | LPC (18:1) | LPC (18:1) |
| 524.370 | LPC (18:0) | LPC (18:0) | ------------ |
| Phosphatidylethanolamine (PE) | |||
| 660.460 | PE (30:1) | 14:0/16:1 | 14:0/16:1 |
| 662.477 | PE (30:0) | 14:0/16:0 | 14:0/16:0 |
| 686.477 | PE (32:2) | 16:1/16:1 | 16:1/16:1 |
| 688.492 | PE (32:1) | 16:0/16:1 | 16:0/16:1 |
| 712.492 | PE (34:3) | 16:0/18:3 and 16:1/18:2 | 16:0/18:3 and 16:1/18:2 |
| 714.508 | PE (34:2) | 16:1/18:1 | 16:1/18:1 |
| 716.524 | PE (34:1) | 16:0/18:1 | 16:0/18:1 |
| 736.492 | PE (36:5) | 16:0/20:5 | 16:0/20:5 |
| 738.508 | PE (36:4) | 16:0/20:4 | ----------- |
| 740.524 | PE (36:3) | ----------- | 18:0/18:3 |
| 742.539 | PE (36:2) | ----------- | 18:1/18:1 and 18:0/18:2 |
| 744.555 | PE (36:1) | ----------- | 18:0/18:1 |
| 786.508 | PE (40:8) | 20:4/20:4 | ----------- |
| Lyso-phosphatidylethanolamine (LPE) | |||
| 450.263 | LPE (16:1) | LPE (16:1) | LPE (16:1) |
| 452.278 | LPE (16:0) | LPE (16:0) | LPE (16:0) |
| 478.294 | LPE (18:1) | LPE (18:1) | LPE (18:1) |
| Phosphatidylglycerol (PG) | |||
| 691.456 | PG (30:1) | ----------- | 14:0/16:1 |
| 717.471 | PG (32:2) | ----------- | 16:0/16:2 and 16:1/16:1 |
| 719.487 | PG (32:1) | 16:1/16:0 and (14:0/18:1) | 16:1/16:0 and 14:0/18:1 |
| 721.502 | PG (32:0) | 16:0/16:0 and 14:0/18:0 | 16:0/16:0 and 14:0/18:0 |
| 745.503 | PG (34:2) | ----------- | 16:1/18:1 and 14:0/18:2 |
| 747.518 | PG (34:1) | 16:0/18:1 and 18:0/16:1 | 16:0/18:1 and 18:0/16:1 |
| 749.534 | PG (34:0) | ----------- | 16:0/18:0 |
| 765.471 | PG (36:6) | 16:1/20:5 and 18:3/18:3 | 16:1/20:5 and 18:3/18:3 |
| 767.487 | PG (36:5) | 16:0/20:5 and 16:1/20:4 | 16:0/20:5 and 16:1/20:4 |
| 769.503 | PG (36:4) | 16:0/20:4 and 18:1/18:3 and 18:2/18:2 | 16:0/20:4 and 18:1/18:3 and 18:2/18:2 |
| 771.518 | PG (36:3) | 18:1/18:2 and 16:0/20:3 | 18:1/18:2 and 16:0/20:3 |
| 773.534 | PG (36:2) | 18:1/18:1 and 16:0/20:2 | 18:1/18:1 and 16:0/20:2 |
| 775.549 | PG (36:1) | 18:0/18:1 | 18:0/18:1 and 16:0/20:1 |
| 801.565 | PG (38:2) | 16:0/22:2 | 16:0/22:2 |
| 803.580 | PG (38:1) | 16:0/22:1 | 16:0/22:1 |
| Lyso-phosphatidylglycerol (LPG) | |||
| 481.257 | LPG (16:1) | LPG (16:1) | LPG (16:1) |
| 483.273 | LPG (16:0) | LPG (16:0) | LPG (16:0) |
| 509.289 | LPG (18:1) | LPG (18:1) | LPG (18:1) |
| Phosphatidic acid (PA) | |||
| 643.434 | PA (32:2)) | 14:0/18:2 and 16:1/16:1 | ----------- |
| 645.450 | PA (32:1) | 14:0/18:1 and 16:0/16:1 | 14:0/18:1 and 16:0/16:1 |
| 647.466 | PA (32:1) | 16:0/16:0 | ----------- |
| 669.450 | PA (34:3) | 16:0/18:3 and 16:1/18:2 14:0/20:3 | ----------- |
| 671.465 | PA (34:2) | 16:0/18:2 | 16:0/18:2 |
| 673.481 | PA (34:1) | 16:1/18:0 and 16:0/18:1 | ----------- |
| 691.434 | PA (36:6) | 16:1/20:5 | ----------- |
| 693.450 | PA (36:5) | 16:0/20:5 | ----------- |
| 695.466 | PA (36:4) | 16:0/20:4 | 16:0/20:4 |
| 697.481 | PA (36:3) | 16:0/20:3 | ----------- |
| 699.497 | PA (36:2) | 16:0/20:2 | 16:0/20:2 |
| 701.513 | PA (36:1) | 16:0/20:1 | 16:0/20:1 |
| Phosphatidylinositol (PI) | |||
| 831.503 | PI (34:3) | ----------- | 16:0/18:3 |
| 833.519 | PI (34:2) | 16:0/18:2 and 16:1/18:1 | 16:0/18:2 and 16:1/18:1 |
| 835.534 | PI (34:1) | 16:0/18:1 | 16:0/18:1 |
| 883.534 | PI (38:5) | ----------- | 18:0/20:5 |
| Inositephosphoceramide (IPC) | |||
| 918.681 | IPC (d44:1) | d18:1/26:0 | ----------- |
| 920.623 | IPC (t42:2-OH) | t18:1/24:1-OH | t18:1/24:1-OH |
| 922.639 | IPC (t42:1-OH) | t18:0/24:1-OH | t18:0/24:1-OH |
| 924.654 | IPC (t42:0-OH) | t18:0/24:0-OH | t18:0/24:0-OH |
| [M + H]+ | Fatty Acyl Composition | ||
|---|---|---|---|
| m/z Theoretical | (C:N) | Blade | Conchocelis |
| 682.562 | DGTS (30:1) | ----------- | 14:0/16:1 |
| 684.578 | DGTS (30:0) | ----------- | 14:0/16:0 |
| 710.593 | DGTS (32:1) | 16:0/16:1 and 14:0/18:1 | 16:0/16:1 and 14:0/18:1 |
| 736.609 | DGTS (34:2) | 16:0/18:2 | 16:0/18:2 |
| 738.625 | DGTS (34:1) | ----------- | 16:0/18:1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Da Costa, E.; Azevedo, V.; Melo, T.; Rego, A.M.; V. Evtuguin, D.; Domingues, P.; Calado, R.; Pereira, R.; Abreu, M.H.; Domingues, M.R. High-Resolution Lipidomics of the Early Life Stages of the Red Seaweed Porphyra dioica. Molecules 2018, 23, 187. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules23010187
Da Costa E, Azevedo V, Melo T, Rego AM, V. Evtuguin D, Domingues P, Calado R, Pereira R, Abreu MH, Domingues MR. High-Resolution Lipidomics of the Early Life Stages of the Red Seaweed Porphyra dioica. Molecules. 2018; 23(1):187. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules23010187
Chicago/Turabian StyleDa Costa, Elisabete, Vitor Azevedo, Tânia Melo, Andreia M. Rego, Dmitry V. Evtuguin, Pedro Domingues, Ricardo Calado, Rui Pereira, Maria H. Abreu, and Maria R. Domingues. 2018. "High-Resolution Lipidomics of the Early Life Stages of the Red Seaweed Porphyra dioica" Molecules 23, no. 1: 187. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules23010187