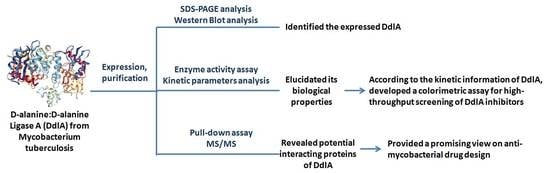
The Biological Properties and Potential Interacting Proteins of d-Alanyl-d-alanine Ligase A from Mycobacterium tuberculosis
Abstract
:1. Introduction
2. Results
2.1. The Detection of Soluble Tb-DdlA Protein Expressed in Escherichia coli
2.2. The Profiles of Tb-DdlA d-Alanyl-d-alanine Ligase Activity
2.2.1. Confirmation of DdlA Activity
2.2.2. Kinetic Analysis of Tb-DdlA
2.2.3. Inhibitory Analysis of Tb-DdlA Using DCS
2.3. The Identification of Tb-DdlA Potential Interaction Partners
3. Discussion
4. Methods
4.1. Strains, Plasmids, and Growth Conditions
4.2. Construction of Tb-ddlA Expression Plasmids
4.3. Expression, Purification, and Identification of Tb-DdlA
4.4. Functional Assay of Tb-DdlA
4.5. Kinetic Analysis of Tb-DdlA
4.6. Colorimetric Inhibition Assay of Tb-DdlA by DCS
4.7. Pull down Assay of Tb-DdlA
4.8. MS/MS Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- WHO. gtbr2017 WHO TB report.pdf. Available online: http://www.who.int/tb/publications/global_report/en/ (accessed on 1 December 2017).
- Zhang, W.; Jones, V.C.; Scherman, M.S.; Mahapatra, S.; Crick, D.; Bhamidi, S.; Xin, Y.; McNeil, M.R.; Ma, Y. Expression, essentiality, and a microtiter plate assay for mycobacterial GlmU, the bifunctional glucosamine-1-phosphate acetyltransferase and N-acetylglucosamine-1-phosphate uridyltransferase. Int. J. Biochem. Cell Biol. 2008, 40, 2560–2571. [Google Scholar] [CrossRef] [PubMed]
- Ba Diallo, A.; Ossoga, G.W.; Daneau, G.; Lo, S.; Ngandolo, R.; Djaibe, C.D.; Djouater, B.; Mboup, S.; de Jong, B.C.; Diallo, A.G.; et al. Emergence and clonal transmission of multi-drug-resistant tuberculosis among patients in Chad. BMC Infect. Dis. 2017, 17, 579. [Google Scholar] [CrossRef] [PubMed]
- Prosser, G.A.; de Carvalho, L.P. Metabolomics Reveal d-alanine: d-alanine Ligase As the Target of d-Cycloserine in Mycobacterium tuberculosis. ACS Med. Chem. Lett. 2013, 4, 1233–1237. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J.; Maurelli, A.T. Characterization of Chlamydia MurC-Ddl, a fusion protein exhibiting D-alanyl-D-alanine ligase activity involved in peptidoglycan synthesis and D-cycloserine sensitivity. Mol. Microbiol. 2005, 57, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Halouska, S.; Fenton, R.J.; Zinniel, D.K.; Marshall, D.D.; Barletta, R.G.; Powers, R. Metabolomics analysis identifies d-alanine-d-alanine ligase as the primary lethal target of d-Cycloserine in mycobacteria. J. Proteom. Res. 2014, 13, 1065–1076. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, Y.; Ebihara, A.; Agari, Y.; Shinkai, A.; Hirotsu, K.; Kuramitsu, S. Structure of D-alanine-D-alanine ligase from Thermus thermophilus HB8: Cumulative conformational change and enzyme-ligand interactions. Acta Crystallogr. Sec. D Biol. Crystallogr. 2009, 65, 1098–1106. [Google Scholar] [CrossRef] [PubMed]
- Bruning, J.B.; Murillo, A.C.; Chacon, O.; Barletta, R.G.; Sacchettini, J.C. Structure of the Mycobacterium tuberculosis d-alanine:d-alanine ligase, a target of the antituberculosis drug D-cycloserine. Antimicrob. Agents Chemother. 2011, 55, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Na, Y.; Song, H.E.; Kim, D.; Park, B.H.; Rho, S.H.; Im, Y.J.; Kim, M.K.; Kang, G.B.; Lee, D.S.; et al. Crystal structure of the apo form of d-alanine: d-alanine ligase (Ddl) from Thermus caldophilus: A basis for the substrate-induced conformational changes. Proteins 2006, 64, 1078–1082. [Google Scholar] [CrossRef] [PubMed]
- Scheffers, D.J.; Pinho, M.G. Bacterial cell wall synthesis: New insights from localization studies. Microbiol. Molecular Biol. Rev. 2005, 69, 585–607. [Google Scholar] [CrossRef] [PubMed]
- Sauvage, E.; Kerff, F.; Terrak, M.; Ayala, J.A.; Charlier, P. The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008, 32, 234–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macheboeuf, P.; Contreras-Martel, C.; Job, V.; Dideberg, O.; Dessen, A. Penicillin binding proteins: Key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol. Rev. 2006, 30, 673–691. [Google Scholar] [CrossRef] [PubMed]
- Pavelka, M.S., Jr.; Jacobs, J.W., Jr. Biosynthesis of diaminopimelate, the precursor of lysine and a component of peptidoglycan, is an essential function of Mycobacterium smegmatis. J. Bacteriol. 1996, 178, 6496–6507. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.M.; Uplekar, S.; Gordon, S.V.; Cole, S.T. A point mutation in cycA partially contributes to the D-cycloserine resistance trait of Mycobacterium bovis BCG vaccine strains. PLoS ONE 2012, 7, e43467. [Google Scholar] [CrossRef] [PubMed]
- Schade, S.; Paulus, W. D-Cycloserine in Neuropsychiatric Diseases: A Systematic Review. Int. J. Neuropsychopharmacol. 2016, 19, pyv102. [Google Scholar] [CrossRef] [PubMed]
- CM, S.; DH, B.; EJ, R. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003, 48, 77–84. [Google Scholar]
- Feng, Z.; Barletta, R.G. Roles of Mycobacterium smegmatis D-Alanine:D-Alanine Ligase and D-Alanine Racemase in the Mechanisms of Action of and Resistance to the Peptidoglycan Inhibitor D-Cycloserine. Antimicrob. Agents Chemother. 2003, 47, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Halouska, S.; Chacon, O.; Fenton, R.J.; Zinniel, D.K.; Barletta, R.G.; Powers, R. Use of NMR metabolomics to analyze the targets of D-cycloserine in mycobacteria: Role of D-alanine racemase. J. Proteom. Res. 2007, 6, 4608–4614. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Samal, S.K.; Routray, S.; Dash, R.; Dixit, A. Identification of oral cancer related candidate genes by integrating protein-protein interactions, gene ontology, pathway analysis and immunohistochemistry. Sci. Rep. 2017, 7, 2472. [Google Scholar] [CrossRef] [PubMed]
- Lampe, J.N. Advances in the Understanding of Protein-Protein Interactions in Drug Metabolizing Enzymes through the Use of Biophysical Techniques. Front. Pharmacol. 2017, 8, 521. [Google Scholar] [CrossRef] [PubMed]
- Modell, A.E.; Blosser, S.L.; Arora, P.S. Systematic Targeting of Protein-Protein Interactions. Trends Pharmacol. Sci. 2016, 37, 702–713. [Google Scholar] [CrossRef] [PubMed]
- Sedan, Y.; Marcu, O.; Lyskov, S.; Schueler-Furman, O. Peptiderive server: Derive peptide inhibitors from protein-protein interactions. Nucleic Acids Res. 2016, 44, W536–W541. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, P.; Miryala, S.; Varshney, U. Use of Mycobacterium smegmatis deficient in ADP-ribosyltransferase as surrogate for Mycobacterium tuberculosis in drug testing and mutation analysis. PLoS ONE 2015, 10, e0122076. [Google Scholar] [CrossRef] [PubMed]
- Valente, W.; Pienaar, E.; Fast, A.; Fluitt, A.; Whitney, S.; Fenton, R.; Barletta, R.; Chacon, O.; Viljoen, H. A Kinetic Study of In Vitro Lysis of Mycobacterium smegmatis. Chem. Eng. Sci. 2009, 64, 1944–1952. [Google Scholar] [CrossRef] [PubMed]
- Wehrmann, A.; Phillipp, B.; Sahm, H.; Eggeling, L. Different modes of diaminopimelate synthesis and their role in cell wall integrity: A study with Corynebacterium glutamicum. J. Bacteriol. 1998, 180, 3159–3165. [Google Scholar] [PubMed]
- McCoy, A.J.; Adams, N.E.; Hudson, A.O.; Gilvarg, C.; Leustek, T.; Maurelli, A.T. l,l-diaminopimelate aminotransferase, a trans-kingdom enzyme shared by Chlamydia and plants for synthesis of diaminopimelate/lysine. Proc. Natl. Acad. Sci. USA 2006, 103, 17909–17914. [Google Scholar] [CrossRef] [PubMed]
- Gokulan, K.; Rupp, B.; Pavelka, M.S., Jr.; Jacobs, W.R., Jr.; Sacchettini, J.C. Crystal structure of Mycobacterium tuberculosis diaminopimelate decarboxylase, an essential enzyme in bacterial lysine biosynthesis. J. Biol. Chem. 2003, 278, 18588–18596. [Google Scholar] [CrossRef] [PubMed]
- Consaul, S.A.; Wright, L.F.; Mahapatra, S.; Crick, D.C.; Pavelka, M.S., Jr. An unusual mutation results in the replacement of diaminopimelate with lanthionine in the peptidoglycan of a mutant strain of Mycobacterium smegmatis. J. Bacteriol. 2005, 187, 1612–1620. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the pCold II-Tb-ddlAplasmid are available from the authors. |







| Reactions | Components | Mean Values of A620nm ± SD | Net Mean ± SD | Z-Factor | |
|---|---|---|---|---|---|
| Reaction 1 | 100 μmol/L ATP; 2 mmol/L d-Ala | 20 μg/mL DdlA | 0.692 ± 0.099 | 0.452 ± 0.099 | 0.187 |
| 100 μmol/L ATP; 2 mmol/L d-Ala | No DdlA | 0.240 ± 0.023 | |||
| Reaction 2 | 75 μmol/L ATP; 2 mmol/L d-Ala | 10 μg/mL DdlA | 0.459 ± 0.013 | 0.291 ± 0.013 | 0.735 |
| 75 μmol/L ATP; 2 mmol/L d-Ala | No DdlA | 0.168 ± 0.013 | |||
| ID | Protein Name | Accession No. (NCBI) | Locus Name/Gene Definition | Protein MW (D) | Protein PI | Pep. Count | Protein Score | Protein Score C. I. % |
|---|---|---|---|---|---|---|---|---|
| A | LuxR family transcriptional regulator | AIU17347.1 | MSMEG_5707, cupin | 12,011.3 | 6.36 | 5 | 285 | 100 |
| B | hypothetical protein LI98_12890 | AIU21016.1 | MSMEG_2589 | 15,412.9 | 6.28 | 7 | 314 | 100 |
| C | elongation factor Tu | YP_885786.1 | MSMEG_1401, tuf | 43,708.6 | 5.18 | 18 | 696 | 100 |
| FAD-dependent oxidoreductase | AIU20161.1 | MSMEG_1682, FMO | 46,185.2 | 5.86 | 13 | 74 | 99.697 | |
| ornithine-oxo-acid transaminase | WP_011727654.1 | MSMEG_1413, rocD | 44,661.1 | 5.62 | 14 | 72 | 99.581 | |
| D | diaminopimelate decarboxylase | YP_889210.1 | MSMEG_4958, lysA | 50,317 | 5.25 | 13 | 224 | 100 |
| cyclopropane-fatty-acyl-phospholipid synthase | YP_890503.1 | MSMEG_6284 | 48,608.7 | 5.99 | 13 | 198 | 100 | |
| carbon-monoxide dehydrogenase large subunit | WP_011727162.1 | MSMEG_0746 | 85,807 | 5.21 | 19 | 60 | 93.366 |
| Strains/Plasmids | Description | Source |
|---|---|---|
| Strains | ||
| E. coli NovaBlue | Used for cloning and propagation of plasmids | Novagen |
| E. coli BL21(DE3) | Used for expressing Tb-DdlA protein | Invitrogen |
| Plasmids | ||
| pMD18-T | Carries ampR gene; used for cloning PCR product with A at 3′ ends | Takara |
| pCold II | Carries ampR gene; contains cold start promoter; used for expressing M. tuberculosis DdlA | Takara |
| pMD18-Tb-ddlA | Carries ampR gene; M. tuberculosis ddlA was cloned to the EcoRV site of pMD18-T | This study |
| pCold II-Tb-ddlA | Carries ampR gene; used for expressing M. tuberculosis DdlA | This study |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.; Xu, Y.; Wang, Y.; Ren, F.; Li, S.; Ding, W.; Ma, Y.; Zhang, W. The Biological Properties and Potential Interacting Proteins of d-Alanyl-d-alanine Ligase A from Mycobacterium tuberculosis. Molecules 2018, 23, 324. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules23020324
Yang S, Xu Y, Wang Y, Ren F, Li S, Ding W, Ma Y, Zhang W. The Biological Properties and Potential Interacting Proteins of d-Alanyl-d-alanine Ligase A from Mycobacterium tuberculosis. Molecules. 2018; 23(2):324. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules23020324
Chicago/Turabian StyleYang, Shufeng, Yuefei Xu, Yan Wang, Feng Ren, Sheng Li, Wenyong Ding, Yufang Ma, and Wenli Zhang. 2018. "The Biological Properties and Potential Interacting Proteins of d-Alanyl-d-alanine Ligase A from Mycobacterium tuberculosis" Molecules 23, no. 2: 324. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules23020324