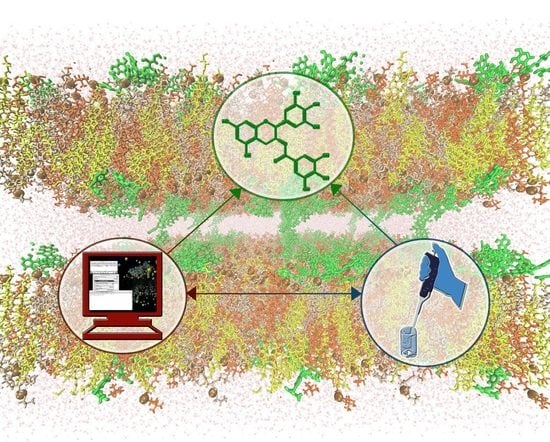
Liposomal Formulations for an Efficient Encapsulation of Epigallocatechin-3-Gallate: An In-Silico/Experimental Approach
Abstract
:1. Introduction
2. Results and Discussion
2.1. Effect of Salts and Lipid Composition on EGCG Encapsulation Efficiency Inside Multilamellar Vesicles (MLVs)
2.2. Interaction of EGCG with Lipidic Bilayer: Simulation Results
2.2.1. Neutral Lipid Models
2.2.2. Anionic Lipid Models
2.2.3. Time-Averaged Distributions of EGCG, Mg2+/Ca2+
3. Materials and Methods
3.1. Computational Methods
3.1.1. Parameterization of Epigallocatechin-3-Gallate and CHEMS
3.1.2. Molecular Dynamics of EGCG and in Mixed Lipid Bilayers
3.1.3. MD Simulation Analyses
3.2. Experimental Materials and Methods
3.2.1. Materials
3.2.2. Preparation of EGCG Liposomes
3.2.3. Physicochemical Characterization of Liposomes
3.2.4. Determination of Encapsulation Efficiency
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Moghimi, S.M.; Hunter, A.C.; Murray, J.C. Nanomedicine: Current status and future prospects. FASEB J. 2005, 19, 311–330. [Google Scholar] [CrossRef] [PubMed]
- Farokhzad, O.C.; Langer, R. Nanomedicine: Developing smarter therapeutic and diagnostic modalities. Adv. Drug Deliv. Rev. 2006, 58, 1456–1459. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chan, J.M.; Gu, F.X.; Rhee, J.W.; Wang, A.Z.; Radovic-Moreno, A.F.; Alexis, F.; Langer, R.; Farokhzad, O.C. Self-Assembled Lipid–Polymer Hybrid Nanoparticles: A Robust Drug Delivery Platform. ACS Nano 2008, 2, 1696–1702. [Google Scholar] [CrossRef] [PubMed]
- Wagner, V.; Dullaart, A.; Bock, A.K.; Zweck, A. The emerging nanomedicine landscape. Nat. Biotechnol. 2006, 24, 1211–1217. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, S.; Eavarone, D.; Capila, I.; Zhao, G.; Watson, N.; Kiziltepe, T.; Sasisekharan, R. Temporal targeting of tumour cells and neovasculature with a nanoscale delivery system. Nature 2005, 436, 568–572. [Google Scholar] [CrossRef] [PubMed]
- Layre, A.; Couvreur, P.; Chacun, H.; Richard, J.; Passirani, C.; Requier, D.; Benoit, J.P.; Gref, R. Novel composite core-shell nanoparticles as busulfan carriers. J. Control Release 2006, 111, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Minnelli, C.; Cianfruglia, L.; Laudadio, E.; Galeazzi, R.; Pisani, M.; Crucianelli, E.; Bizzaro, D.; Armeni, T.; Mobbili, G. Selective induction of apoptosis in MCF7 cancer-cell by targeted liposomes functionalised with mannose-6-phosphate. J. Drug Target. 2017, 26, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Crucianelli, E.; Bruni, P.; Frontini, A.; Massaccesi, L.; Pisani, M.; Smorlesi, A.; Mobbili, G. Liposomes containing mannose-6-phosphate-cholesteryl conjugates for lysosome-specific delivery. RSC Adv. 2014, 4, 58204–58207. [Google Scholar] [CrossRef]
- Galeazzi, R.; Bruni, P.; Crucianelli, E.; Laudadio, E.; Marini, M.; Massaccesi, L.; Mobbili, G.; Pisani, M. Liposome-based gene delivery systems containing a steroid derivative: Computational and small angle X-ray diffraction study. RSC Adv. 2015, 5, 54070–54078. [Google Scholar] [CrossRef]
- Torchilin, V.P. Recent advances with liposomes as pharmaceutical carriers. Nat. Rev. Drug Discov. 2005, 4, 145–160. [Google Scholar]
- Katiyar, S.K.; Ahmad, N.; Mukhtar, H. Green tea and skin. Arch. Dermatol. 2000, 136, 989–994. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Bachrach, U. The specific anti-cancer activity of green tea (-)-epigallocatechin-3-gallate (EGCG). Amino Acids 2002, 22, 131–143. [Google Scholar]
- Deng, Y.T.; Lin, J.K. EGCG Inhibits the Invasion of Highly Invasive CL1-5 Lung Cancer Cells through Suppressing MMP-2 Expression via JNK Signaling and Induces G2/M Arrest. J. Agric. Food Chem. 2011, 59, 13318–13327. [Google Scholar] [CrossRef] [PubMed]
- Nakachi, K.; Matsuyama, S.; Miyake, S.; Suganuma, M.; Imai, K. Preventive effects of drinking green tea on cancer and cardiovascular disease: Epidemiological evidence for multiple targeting prevention. Biofactors 2000, 13, 49–54. [Google Scholar] [CrossRef]
- Yang, C.S.; Maliakal, P.; Meng, X. Inhibition of carcinogenesis by tea. Annu. Rev. Pharmacol. Toxicol. 2002, 42, 25–54. [Google Scholar] [CrossRef] [PubMed]
- Nieman, D.C.; Henson, D.A.; Maxwell, K.R.; Williams, A.S.; McAnulty, S.R.; Jin, F.; Shanely, R.A.; Lines, T.C. Effects of quercetin and EGCG on mitochondrial biogenesis and immunity. Med. Sci. Sports Exerc. 2009, 41, 1467–1475. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.H.; Ho, M.L.; Chang, J.K.; Hung, S.H.; Wang, G.J. Green tea catechin enhances osteogenesis in a bone marrow mesenchymal stem cell line. Osteoporos. Int. 2005, 16, 2039–2045. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zheng, Y.; Chow, M.S.; Zuo, Z. Investigation of intestinal absorption and disposition of green tea catechins by Caco-2 monolayer model. Int. J. Pharm. 2004, 287, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Granja, A.; Pinheiro, M.; Reis, S. Epigallocatechin gallate nanodelivery systems for cancer therapy. Nutrients 2016, 8, 307. [Google Scholar] [CrossRef]
- Daeihamed, M.; Dadashzadeh, S.; Haeri, A.; Akhlaghi, M.F. Potential of Liposomes for Enhancement of Oral Drug Absorption. Curr. Drug Deliv. 2017, 14, 289–303. [Google Scholar] [CrossRef] [PubMed]
- Sang, S.; Lambert, J.D.; Ho, C.T.; Yang, C.S. The chemistry and biotransformation of tea constituents. Pharmacol. Res. 2011, 64, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Martın-Molina, A.; Rodrıguez-Beas, C.; Faraudo, J. Effect of Calcium and Magnesium on Phosphatidylserine Membranes: Experiments and All-Atomic Simulations. Biophys. J. 2012, 102, 2095–2103. [Google Scholar] [CrossRef] [PubMed]
- Binder, H.; Zschörnig, O. The effect of metal cations on the phase behavior and hydration characteristics of phospholipid membranes. Chem. Phys. Lipids 2002, 115, 39–61. [Google Scholar] [CrossRef]
- Ohki, S.; Arnold, K. A mechanism for ion-induced lipid vesicle fusion. Colloids Surf. B 2000, 18, 83–97. [Google Scholar] [CrossRef]
- Boettcher, J.M.; Davis-Harrison, R.L.; Clay, M.C.; Nieuwkoop, A.J.; Ohkubo, Y.Z.; Tajkhorshid, E.; Morrissey, J. H.; Rienstra, C.M. Atomic view of Calcium-Induced Clustering of Phosphatidylserine in Mixed Lipid Bilayers. Biochemistry 2011, 50, 2264–2273. [Google Scholar] [CrossRef] [PubMed]
- Laudadio, E.; Mobbili, G.; Minnelli, C.; Massaccesi, L.; Galeazzi, R. Salts Influence Cathechins and Flavonoids Encapsulation in Liposomes: A Molecular Dynamics Investigation. Mol. Inform. 2017, 36, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Briuglia, M.L.; Rotella, C.; McFarlane, A.; Lamprou, D.A. Influence of cholesterol on liposome stability and on in vitro drug release. Drug Deliv. Transl. Res. 2015, 5, 231–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakayama, T.; Hashimoto, T.; Kajiya, K.; Kumazawa, S. Affinity of polyphenols for lipid bilayers. Biofactors 2000, 13, 147–151. [Google Scholar] [CrossRef]
- Alvarez, M.S.; Esperança, J.; Deive, J.F.; Sanromán, M.A.; Rodriguez, A. A biocompatible stepping stone for the removal of emerging contaminants. Sep. Purif. Technol. 2015, 153, 91–98. [Google Scholar] [CrossRef]
- Lanigan, R.S.; Yamarik, T.A. Cosmetic Ingredient Review. Expert panel. Final report on the safety assessment of sorbitan caprylate, sorbitan cocoate, sorbitan diisostearate, sorbitan dioleate, sorbitan distearate, sorbitan isostearate, sorbitan olivate, sorbitan sesquiisostearate, sorbitan sesquistearate, and sorbitan triisostearate. Int. J. Toxicol. 2002, 21, 93–112. [Google Scholar] [PubMed]
- Juárez-Osornio, C.; Gracia-Fadrique, J. Structures similar to lipid emulsions and liposomes. Dipalmitoylphosphatidylcholine, Cholesterol, Tween 20- Span 20 or Tween 80-Span 80 in aqueous media. J. Liposome Res. 2017, 27, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Yang, L.; Zhang, Q.; Jiang, H.B.; Jang, H. Effects of ion interactions with a cholesterol-rich bilayer. Biochem. Biophys. Res. Comm. 2015, 468, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Du, Y.; Cang, X.; Wang, J.; Chen, Z.; Yang, H.; Jiang, H. Binding competition to the POPG lipid bilayer of Ca2+, Mg2+, Na+, and K+ in different ion mixtures and biological implication. J Phys. Chem. B 2013, 117, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Migalska-Zalas, A.; Kityk, I.V.; Bakasse, M.; Sahraoui, B. Features of the alkynyl ruthenium chromophore with modified anionic subsystem UV absorption. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2008, 69, 178–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodman, J.M.; James, J.J.; Whiting, A. MM2 force field parameters for compounds containing the diazoketone function. J. Chem. Soc. Perkin Trans. 2 1994, 2, 109–116. [Google Scholar] [CrossRef]
- Onufriev, A.V.; Sigalov, G. A strategy for reducing gross errors in the generalized Born models of implicit solvation. J. Chem. Phys. 2011, 134, 164104. [Google Scholar] [CrossRef] [PubMed]
- Galeazzi, R. Molecular Dynamics as a Tool in Rational Drug Design: Current Status and Some Major Applications. Curr. Comput-Aid Drug Des. 2009, 5, 225–240. [Google Scholar] [CrossRef]
- Huang, J.; MacKerell, A.D., Jr. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef] [PubMed]
- Eastwood, J.W.; Hockney, R.W.; Lawrence, D.N. P3M3DP—The three-dimensional periodic particle-particle/particle-mesh program. Comput. Phys. Commun. 1980, 19, 161–272. [Google Scholar] [CrossRef]
- Galeazzi, R.; Martelli, G.; Marcucci, E.; Mobbili, G.; Natali, D.; Orena, M.; Rinaldi, S. A New Conformationally Restricted Mimetic of Dipeptide EG—Synthesis of an Analogue of FEG. Eur. J. Org. Chem. 2007, 26, 4402–4407. [Google Scholar] [CrossRef]
- Galeazzi, R.; Martelli, G.; Marcucci, E.; Orena, M.; Rinaldi, S.; Lattanzi, R.; Negri, L. Analogues of both Leu- and Met-enkephalin containing a constrained dipeptide isostere prepared from a Baylis-Hillman adduct. Amino Acids 2010, 38, 1057–1065. [Google Scholar] [CrossRef] [PubMed]
- Galeazzi, R.; Massaccesi, L.; Piva, F.; Principato, G.; Laudadio, E. Insights into the influence of 5-HT2c aminoacidic variants with the inhibitory action of serotonin inverse agonists and antagonists. J. Mol. Model. 2014, 20, 2120. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 09, Revision D.01; Gaussian, Inc.: Wallingford CT, USA, 2016.
- Raghavachari, K. Perspective on “Density functional thermochemistry. III. The role of exact exchange. Theor. Chem. Acc. 2000, 103, 361–363. [Google Scholar] [CrossRef]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B Condens. Matter. 1988, 37, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Vosko, S.H.; Wilk, L.; Nusair, M. Accurate spin-dependent electron liquid correlation energies for local spin density calculations: A critical analysis. Can. J. Phys. 1980, 58, 80–159. [Google Scholar] [CrossRef]
- Stephens, P.J.; Devlin, F.J.; Chabalowski, C.F.; Frisch, M.J. Ab Initio Calculation of Vibrational Absorption and Circular Dichroism Spectra Using Density Functional Force Fields: A Comparison of Local, Nonlocal, and Hybrid Density Functionals. J. Phys. Chem. 1994, 98, 11623–11627. [Google Scholar] [CrossRef]
- Becke, A.D. Densityfunctional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- Galeazzi, R.; Martelli, G.; Orena, M.; Rinaldi, S. Stereoselective alkylation of chiral pyrrolidin-2-ones leading to novel conformationally restricted analogues of 3-methylaspartic acid: A computational investigation. Monatsh. Chem. 2012, 143, 1397–1403. [Google Scholar] [CrossRef]
- Mayne, C.G.; Saam, J.; Schulten, K.; Tajkhorshid, E.; Gumbart, J.C. Rapid parameterization of small molecules using the Force Field Toolkit. J. Comput. Chem. 2013, 34, 2757–2770. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Cheatham, T.E., 3rd; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M., Jr.; Onufriev, A.; Simmerling, C.; Wang, B.R.; Woods, J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [PubMed]
- Weiner, S.; Kollman, P.A.; Case, D.A.; Singh, U.C.; Ghio, C.; Alagona, G.; Weiner, S.P., Jr.; Weiner, P. A new force field for molecular mechanical simulation of nucleic acids and proteins. J. Am. Chem. Soc. 1984, 106, 765–784. [Google Scholar] [CrossRef]
- Klauda, J.B.; Venable, R.M.; Freites, J.A.; O’Connor, J.W.; Tobias, D.J.; Mondragon-Ramirez, C.; Vorobyov, I.; MacKerell, A.D., Jr.; Pastor, R.W. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. J. Phys. Chem. B 2010, 114, 7830–7843. [Google Scholar] [CrossRef] [PubMed]
- Domanski, J.; Stansfeld, P.J.; Sansom, M.S.; Beckstein, O. Lipidbook: A public repository for force-field parametersused in membrane simulations. J. Membr. Biol. 2010, 236, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Soper, A.K.; Bruni, F.; Ricci, M.A. Site-site pair correlation functions of water from 25 to 400 °C: Revised analysis of new and old refraction data. J. Chem. Phys. 1997, 106, 247–254. [Google Scholar] [CrossRef]
- Villanueva, D.Y.; Lim, J.B.; Klauda, J.B. Influence of Ester-Modified Lipids on Bilayer Structure. Langmuir 2013, 29, 14196–14203. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Rui, H.; Lim, J.B.; Klauda, J.B.; Im, W. Cholesterol Flip-Flop: Insights from Free Energy Simulation Studies. J. Phys. Chem. B 2010, 114, 13342–13348. [Google Scholar] [CrossRef] [PubMed]
- Klauda, J.B.; Brooks, B.R.; Pastor, R.W. Dynamical motions of lipids and a finite size effect in simulations of bilayers. J. Chem. Phys. 2006, 125, 144710. [Google Scholar] [CrossRef] [PubMed]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Ewald, P.P. The calculation of optical and electrostatic grid potential. Ann. Phys. 1921, 64, 253–287. [Google Scholar] [CrossRef]
- Hockney, R.W.; Eastwood, J.W. Computer Experiments using Particle Models. In Computer Simulation Using Particles, 1st ed.; McGraw-Hill: New York, NY, USA, 1988. [Google Scholar]
- Pronk, S.; Pall, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; van der Spoel, D.; et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Porasso, R.D.; Cascales, J.J.L. A criterion to identify the equilibration time in lipid bilayer simulations. Pap. Phys. 2012, 4, 040005. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Melcrová, A.; Pokorna, S.; Pullanchery, S.; Kohagen, M.; Jurkiewicz, P.; Hof, M.; Jungwirth, P.; Cremer, P.S.; Cwiklik, L. The complex nature of calcium cation interactions with phospholipid bilayers. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Szoka, F., Jr.; Papahadjopoulos, D. Procedure for preparation of liposomes with large internal aqueous space and high capture by reverse-phase evaporation. Proc. Natl. Acad. Sci. USA 1978, 75, 4194–4198. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Xu, S.; Xia, S.; Zhang, X. Effect of different preparation methods on physico-chemical properties of salidroside liposomes. J. Agric. Food Chem. 2007, 55, 3089–3095. [Google Scholar] [CrossRef] [PubMed]
- Pignatello, R.; Nicolosi, D.; Nicolosi, V.M. Fusogenic liposomes as new carriers to enlarge the spectrum of action of antibiotic drugs against Gram-negative bacteria. In Science against Microbial Pathogens: Communicating Current Research and Technological Advances, 1st ed.; Formatex Research Center: Bajadoz, Spain, 2008. [Google Scholar]
- Price, W.E.; Spitzer, J.C. The kinetics of extraction of individual flavanols and caffeine from a Japanese green tea (Sen Cha Uji Tsuyu) as a function of temperature. Food Chem. 1994, 50, 19–23. [Google Scholar] [CrossRef]
- Stewart, J.C. Colorimetric determination of phospholipids with ammonium ferrothiocyanate. Anal Biochem. 1980, 104, 10–14. [Google Scholar] [CrossRef]
- Onyesom, I.; Lamprou, D.A.; Sygellou, L.; Owusu-Ware, S.K.; Antonijevic, M.; Chowdhry, B.Z.; Douroumis, D. Sirolimus Encapsulated Liposomes for Cancer Therapy: Physicochemical and Mechanical Characterization of Sirolimus Distribution within Liposome Bilayers. Mol. Pharm. 2013, 10, 4281–4293. [Google Scholar] [CrossRef] [PubMed]
- Kajiya, K.; Kumazawa, S.; Nakayama, T. Steric effects on interaction of tea catechins with lipid bilayers. Biosci. Biotechnol. Biochem. 2001, 65, 2638–2643. [Google Scholar] [CrossRef] [PubMed]
- Castillo Cohen de Pace, R.; Liu, X.; Sun, M.; Nie, S.; Zhang, J.; Cai, Q.; Gao, W.; Pan, X.; Fan, Z.; Wang, S. Anticancer activities of (-)-epigallocatechin-3-gallate encapsulated nanoliposomes in MCF7 breast cancer cells an anticancer effect of EGCG. J. Liposome Res. 2013, 23, 187–196. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |









| Liposomes Formulation | Molar Ratio Mg2+/EGCG | Encapsulation Efficiency (%) ± SD | Molar Ratio Ca2+/EGCG | Encapsulation Efficiency (%) ± SD |
|---|---|---|---|---|
| 0 | 53.5 ± 6.2 | 0 | 53.5 ± 6.2 | |
| 1:1 | 71.7 ± 7.8 | 1:1 | 32.7 ± 4.3 | |
| Neutral system | 3:1 | 79.1 ± 5.3 | 3:1 | 23.9 ± 2.4 |
| 5:1 | 82.3 ± 3.2 | 5:1 | 12.6 ± 6.4 | |
| 6:1 | 79.2 ± 3.5 | 6:1 | 4.1 ± 1.9 | |
| 0 | 65.5 ± 3.8 | 0 | 65.5 ± 3.8 | |
| 1:1 | 76.7 ± 5.5 | 1:1 | 9.3 ± 2.5 | |
| Anionic system | 3:1 | 82.6 ± 2.3 | 3:1 | 5.9 ± 3.6 |
| 5:1 | 98.9 ± 2.6 | 5:1 | 14.7 ± 1.7 | |
| 6:1 | 76.8 ± 4.2 | 6:1 | 9.9 ± 5.3 |
| Neutral Model | Areapl | Membrane Thickness |
| No divalent sals | 0.83 ± 0.01 nm2 | 4.13 ± 0.43 nm |
| 1:1 CaCl2: EGCG | 0.83 ± 0.04 nm2 | 4.01 ± 0.42 nm |
| 1:1 MgCl2: EGCG | 0.81 ± 0.02 nm2 | 4.22 ± 0.39 nm |
| 3:1 CaCl2: EGCG | 0.79 ± 0.06 nm2 | 3.98 ± 0.41 nm |
| 3:1 MgCl2: EGCG | 0.84 ± 0.03 nm2 | 4.29 ± 0.44 nm |
| 5:1 CaCl2: EGCG | 0.81 ± 0.04 nm2 | 3.79 ± 0.42 nm |
| 5:1 MgCl2: EGCG | 0.87 ± 0.03 nm2 | 4.31 ± 0.38 nm |
| 6:1 CaCl2: EGCG | 0.81 ± 0.05 nm2 | 3.78 ± 0.44 nm |
| 6:1 MgCl2: EGCG | 0.82 ± 0.02 nm2 | 4.30 ± 0.37 nm |
| Anionic Model | Areapl | Membrane Thickness |
| No divalent sals | 0.85 ± 0.03 nm2 | 4.12 ± 0.36 nm |
| 1:1 CaCl2: EGCG | 0.72 ± 0.02 nm2 | 3.70 ± 0.47 nm |
| 1:1 MgCl2: EGCG | 0.91 ± 0.02 nm2 | 4.33 ± 0.37 nm |
| 3:1 CaCl2: EGCG | 0.75 ± 0.05 nm2 | 3.73 ± 0.48 nm |
| 3:1 MgCl2: EGCG | 0.94 ± 0.03 nm2 | 4.42 ± 0.36 nm |
| 5:1 CaCl2: EGCG | 0.77 ± 0.03 nm2 | 3.61 ± 0.46 nm |
| 5:1 MgCl2: EGCG | 1.16 ± 0.04 nm2 | 4.79 ± 0.32 nm |
| 6:1 CaCl2: EGCG | 0.78 ± 0.05 nm2 | 3.60 ± 0.47 nm |
| 6:1 MgCl2:EGCG | 0.91 ± 0.03 nm2 | 4.21 ± 0.35 nm |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laudadio, E.; Minnelli, C.; Amici, A.; Massaccesi, L.; Mobbili, G.; Galeazzi, R. Liposomal Formulations for an Efficient Encapsulation of Epigallocatechin-3-Gallate: An In-Silico/Experimental Approach. Molecules 2018, 23, 441. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules23020441
Laudadio E, Minnelli C, Amici A, Massaccesi L, Mobbili G, Galeazzi R. Liposomal Formulations for an Efficient Encapsulation of Epigallocatechin-3-Gallate: An In-Silico/Experimental Approach. Molecules. 2018; 23(2):441. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules23020441
Chicago/Turabian StyleLaudadio, Emiliano, Cristina Minnelli, Adolfo Amici, Luca Massaccesi, Giovanna Mobbili, and Roberta Galeazzi. 2018. "Liposomal Formulations for an Efficient Encapsulation of Epigallocatechin-3-Gallate: An In-Silico/Experimental Approach" Molecules 23, no. 2: 441. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules23020441