Comparative Evaluation of the Antioxidant and Anti-Alzheimer’s Disease Potential of Coumestrol and Puerarol Isolated from Pueraria lobata Using Molecular Modeling Studies
Abstract
:1. Introduction
2. Results
2.1. DPPH and ONOO− Scavenging Potentials of Coumestrol and Puerarol
2.2. ONOO−-Mediated Western Blot Analysis of Coumestrol and Puerarol
2.3. Anti-AD Potential of Coumestrol and Puerarol
2.4. Enzyme Kinetic Analysis of Coumestrol and Puerarol for ChEs and BACE1
2.5. Molecular Docking Simulation Studies of Coumestrol on ChEs
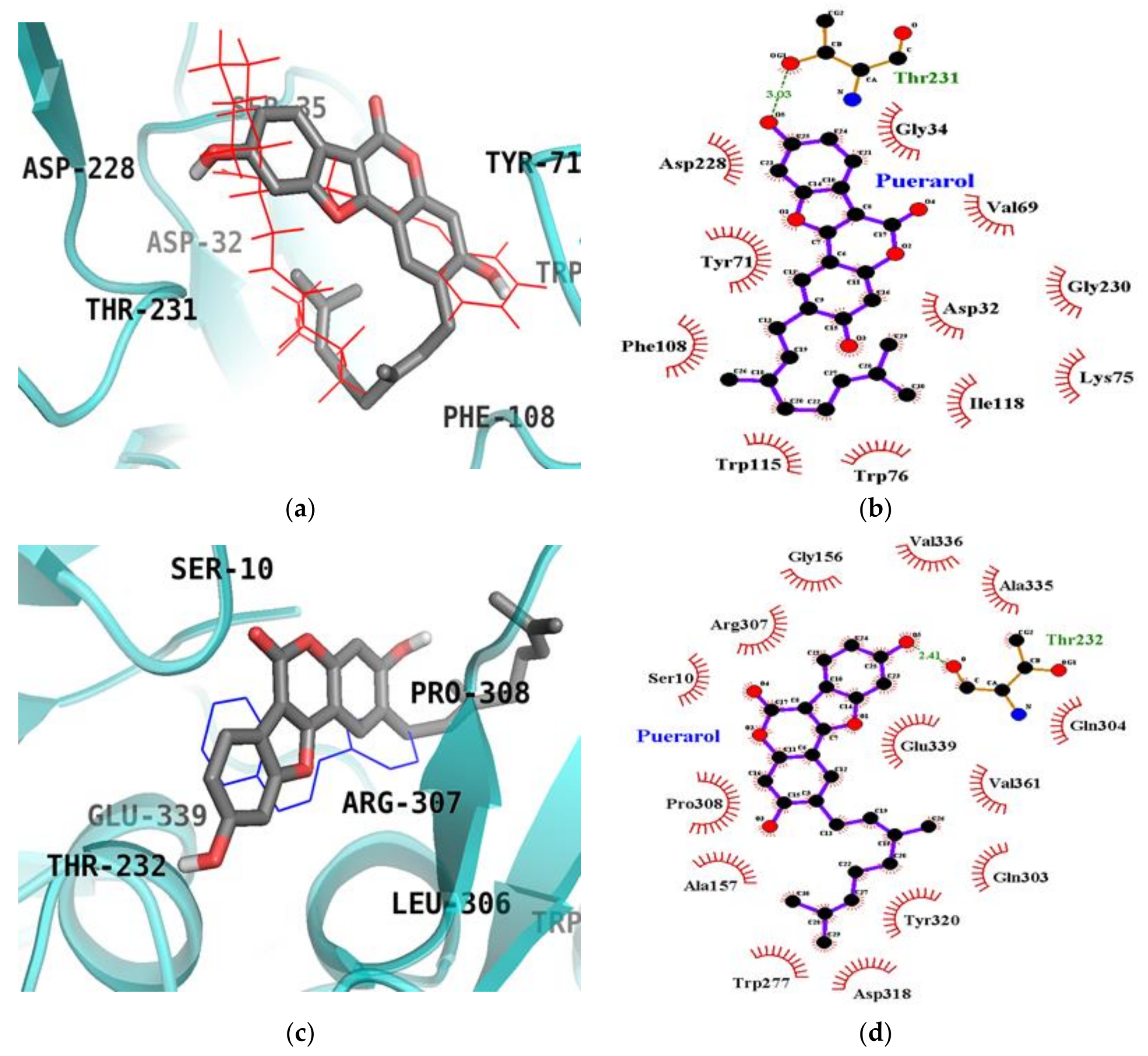
2.6. Molecular Docking Simulation Studies of Puerarol on BACE1
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. DPPH Radical Scavenging Activity
4.3. Assay of ONOO− Scavenging Activity
4.4. Western Blot Analysis for Inhibition of ONOO−-Mediated Nitrotyrosine Formation
4.5. In Vitro ChE Enzyme Assay
4.6. In Vitro BACE1 Enzyme Assay
4.7. Kinetic Parameters for AChE, BChE, and BACE1 Inhibition
4.8. ChEs and BACE1 Molecular Docking Simulations
4.9. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Area-Gomez, E.; Schon, E.A. Alzheimer Disease. Adv. Exp. Med. Biol. 2017, 997, 149–156. [Google Scholar] [PubMed]
- Roher, A.E.; Kokjohn, T.A.; Clarke, S.G.; Sierks, M.R.; Maarouf, C.L.; Serrano, G.E.; Sabbagh, M.S.; Beach, T.G. APP/Aβ structural diversity and Alzheimer’s disease pathogenesis. Neurochem. Int. 2017, 110, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Vassar, R. BACE1 inhibitor drugs in clinical trials for Alzheimer’s disease. Alzheimers Res. Ther. 2014, 6, 89. [Google Scholar] [CrossRef] [PubMed]
- Francis, P.T.; Palmer, A.M.; Snape, M.; Wilcock, G.K. The cholinergic hypothesis of Alzheimer’s disease: A review of progress. J. Neurol. Neurosurg. Psychiatry 1999, 66, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Rao, A.A.; Sridhar, G.R.; Das, U.N. Elevated butyrylcholinesterase and acetylcholinesterase may predict the development of type 2 diabetes mellitus and Alzheimer’s disease. Med. Hypotheses 2007, 69, 1272–1276. [Google Scholar] [CrossRef]
- Thapa, A.; Carroll, N.J. Dietary Modulation of Oxidative Stress in Alzheimer’s Disease. Int. J. Mol. Sci. 2017, 18, 1583. [Google Scholar] [CrossRef] [PubMed]
- Swomley, A.M.; Förster, S.; Keeney, J.T.; Triplett, J.; Zhang, Z.; Sultana, R.; Butterfield, D.A. Abeta, oxidative stress in Alzheimer disease: Evidence based on proteomics studies. Biochim. Biophys. Acta-Mol. Basis Dis. 2014, 1842, 1248–1257. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.L.; Li, Q.X.; Ciccotosto, G.D.; Crouch, P.J.; Culvenor, J.G.; White, A.R.; Evin, G. Mild oxidative stress induces redistribution of BACE1 in non-apoptotic conditions and promotes the amyloidogenic processing of Alzheimer’s disease amyloid precursor protein. PLoS ONE 2013, 8, e61246. [Google Scholar] [CrossRef]
- Feng, Y.; Wang, X. Antioxidant therapies for Alzheimer’s disease. Oxid. Med. Cell. Longev. 2012, 2012. [Google Scholar] [CrossRef]
- Thompson, S.; Lanctôt, K.L.; Herrmann, N. The benefits and risks associated with cholinesterase inhibitor therapy in Alzheimer’s disease. Expert Opin. Drug Saf. 2004, 3, 425–440. [Google Scholar] [CrossRef] [PubMed]
- Hung, S.Y.; Fu, W.M. Drug candidates in clinical trials for Alzheimer’s disease. Int. J. Biomed. Sci. 2017, 24, 47. [Google Scholar] [CrossRef] [PubMed]
- Rasul, A.; Millimouno, F.M.; Ali Eltayb, W.; Ali, M.; Li, J.; Li, X. Pinocembrin: A novel natural compound with versatile pharmacological and biological activities. Biomed. Res. Int. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Bedell, S.; Nachtigall, M.; Naftolin, F. The pros and cons of plant estrogens for menopause. J. Steroid Biochem. Mol. Biol. 2014, 139, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Bickoff, E.M.; Booth, A.N.; Lyman, R.L.; Livingston, A.L.; Thompson, C.R.; Kohler, G.O. Plant estrogens, isolation of a new estrogen from ladino clover. J. Agric. Food Chem. 1958, 6, 536–539. [Google Scholar] [CrossRef]
- Bickoff, E.M.; Spencerm, R.R.; Witt, S.C.; Knuckles, B.E. Studies on the Chemical and Biological Properties of Coumestrol and Related Compounds; US Department of Agriculture: Washington, DC, USA, 1969; Volume 1408, pp. 1–83. [Google Scholar]
- Castro, C.C.; Pagnussat, A.S.; Orlandi, L.; Worm, P.; Moura, N.; Etgen, A.M.; Netto, C.A. Coumestrol has neuroprotective effects before and after global cerebral ischemia in female rats. Brain Res. 2012, 1474, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Humfrey, C.D. Phytoestrogens and human health effects: Weighing up the current evidence. Nat. Toxins 1998, 6, 51–59. [Google Scholar] [CrossRef]
- Park, G.; Baek, S.; Kim, J.E.; Lim, T.G.; Lee, C.C.; Yang, H.; Kang, Y.G.; Park, J.S.; Augustin, M.; Mrosek, M.; et al. Flt3 is a target of coumestrol in protecting against UVB-induced skin photoaging. Biochem. Pharmacol. 2015, 98, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Chandsawangbhuwana, C.; Baker, M.E. 3D models of human ERα and ERβ complexed with coumestrol. Steroids 2014, 80, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.J.; Son, H.J.; Ahn, J.; Jung, C.H.; Ha, T. Coumestrol modulates Akt and Wnt/β-catenin signaling during the attenuation of adipogenesis. Food Funct. 2016, 7, 4984–4991. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.A.; Park, N.H.; Na, Y.J.; Lee, H.K.; Lee, J.H.; Kim, Y.J.; Lee, C.S. Coumestrol down-regulates melanin production in Melan—A murine melanocytes through degradation of tyrosinase. Biol. Pharm. Bull. 2017, 40, 535–539. [Google Scholar] [CrossRef] [PubMed]
- Koirala, P.; Seong, S.H.; Jung, H.A.; Choi, J.S. Comparative molecular docking studies of lupeol and lupenone isolated from Pueraria lobata that inhibits BACE1: Probable remedies for Alzheimer’s disease. Asian Pac. J. Trop. Med. 2017, 10, 1117–1122. [Google Scholar] [CrossRef] [PubMed]
- Seong, S.H.; Roy, A.; Jung, H.A.; Jung, H.J.; Choi, J.S. Protein tyrosine phosphatase 1B and α-glucosidase inhibitory activities of Pueraria lobata root and its constituents. J. Ethnopharmacol. 2016, 194, 706–716. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Heo, J.H.; Kim, Y.S.; Kang, S.S.; Choi, J.S.; Lee, S.M. Protective effect of daidzin against d-galactosamine and lipopolysaccharide-induced hepatic failure in mice. Phytother. Res. 2009, 23, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Mangialasche, F.; Polidori, M.C.; Monastero, R.; Ercolani, S.; Camarda, C.; Cecchetti, R.; Mecocci, P. Biomarkers of oxidative and nitrosative damage in Alzheimer’s disease and mild cognitive impairment. Ageing Res. Rev. 2009, 8, 285–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwak, Y.D.; Wang, R.; Li, J.J.; Zhang, Y.W.; Xu, H.; Liao, F.F. Differential regulation of BACE1 expression by oxidative and nitrosative signals. Mol. Neurodegener. 2011, 6, 17. [Google Scholar] [CrossRef] [PubMed]
- Getchell, M.L.; Shah, D.S.; Buch, S.K.; Davis, D.G.; Getchell, T.V. 3-Nitrotyrosine immunoreactivity in olfactory receptor neurons of patients with Alzheimer’s disease: Implications for impaired odor sensitivity. Neurobiol. Aging 2003, 4, 663–673. [Google Scholar] [CrossRef]
- Butterfield, D.A.; Drake, J.; Pocernich, C.; Castegna, A. Evidence of oxidative damage in Alzheimer’s disease brain: Central role for amyloid β-peptide. Trends Mol. Med. 2001, 7, 548–554. [Google Scholar] [CrossRef]
- Booth, N.L.; Overk, C.R.; Yao, P.; Burdette, J.E.; Nikolic, D.; Chen, S.N.; Bolton, J.L.; van Breemen, R.B.; Pauli, G.F.; Farnsworth, N.R. The chemical and biologic profile of a red clover (Trifolium pratense L.) phase II clinical extract. J. Altern. Complement. Med. 2006, 12, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Jeon, H.Y.; Seo, D.B.; Shin, H.J.; Lee, S.J. Effect of Aspergillus oryzae-challenged germination on soybean isoflavone content and antioxidant activity. J. Agric. Food Chem. 2012, 60, 2807–2814. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, T.; Liu, J.; Liu, Y.; Zhang, J.; Lin, J.; Zhao, Z.; Chen, D.F. Effect and mechanism of wedelolactone as antioxidant-coumestan on OH-treated mesenchymal stem cells. Arabian J. Chem. 2017. [Google Scholar] [CrossRef]
- Isoda, H.; Talorete, T.P.; Kimura, M.; Maekawa, T.; Inamori, Y.; Nakajima, N.; Seki, H. Phytoestrogens genistein and daidzin enhance the acetylcholinesterase activity of the rat pheochromocytoma cell line PC12 by binding to the estrogen receptor. Cytotechnology 2002, 40, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Ramasamy, K.; Majeed, A.B.; Mani, V. Enhancement of β-secretase inhibition and antioxidant activities of tempeh, a fermented soybean cake through enrichment of bioactive aglycones. Pharm. Biol. 2015, 53, 758–766. [Google Scholar] [CrossRef] [PubMed]
- Balkis, A.; Tran, K.; Lee, Y.Z.; Ng, K. Screening flavonoids for inhibition of acetylcholinesterase identified baicalein as the most potent inhibitor. J. Agric. Sci. 2015, 7, 26–35. [Google Scholar] [CrossRef]
- Zafar, A.; Ahmad, S.; Naseem, I. Insight into the structural stability of coumestrol with human estrogen receptor α and β subtypes: A combined approach involving docking and molecular dynamics simulation studies. RSC Adv. 2015, 5, 81295–81312. [Google Scholar] [CrossRef]
- Blois, M.S. Antioxidant determination by the use of a stable free radical. Nature 1958, 181, 1199–1200. [Google Scholar] [CrossRef]
- Kooy, N.W.; Royall, J.A.; Ischiropoulos, H.; Beckman, J.S. Peroxynitrite-mediated oxidation of dihydrorhodamine 123. Free Radic. Biol. Med. 1994, 16, 149–156. [Google Scholar] [CrossRef]
- Aulak, K.S.; Miyagi, M.; Yan, L.; West, K.A.; Massillon, D.; Crabb, J.W.; Stuehr, D.J. Proteomic method identifies proteins nitrated in vivo during inflammatory challenge. Proc. Natl. Acad. Sci. USA 2001, 98, 12056–12061. [Google Scholar] [CrossRef] [PubMed]
- Ellman, G.L.; Courtney, K.D.; Andres, V.; Featherstone, R.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961, 7, 88–95. [Google Scholar] [CrossRef]
- Jung, H.A.; Jin, S.E.; Choi, R.J.; Kim, D.H.; Kim, Y.S.; Ryu, J.H.; Kim, D.W.; Son, Y.K.; Park, J.J.; Choi, J.S. Anti-amnesic activity of neferine with antioxidant and anti-inflammatory capacities, as well as inhibition of ChEs and BACE1. Life Sci. 2010, 87, 420–430. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Shi, A.; He, F.; Li, X. Synthesis, biological evaluation, and molecular modeling of berberine derivatives as potent acetylcholinesterase inhibitors. Bioorg. Med. Chem. 2010, 18, 1244–1251. [Google Scholar] [CrossRef] [PubMed]
- Shimmyo, Y.; Kihara, T.; Akaike, A.; Niidome, T.; Sugimoto, H. Flavonols and flavones as BACE-1 inhibitors: Structure-activity relationship in cell-free, cell-based and in silico studies reveal novel pharmacophore features. Biochim. Biophys. Acta 2008, 1780, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Lineweaver, H.; Burk, D. The determination of enzyme dissociation constants. J. Am. Chem. Soc. 1934, 56, 658–666. [Google Scholar] [CrossRef]
- Harel, M.; Schalk, I.; Ehret-Sabatier, L.; Bouet, F.; Goeldner, M.; Hirth, C.; Axelsen, P.H.; Silman, I.; Sussman, J.L. Quaternary ligand binding to aromatic residues in the active-site gorge of acetylcholinesterase. Proc. Natl. Acad. Sci. USA 1993, 90, 9031–9035. [Google Scholar] [CrossRef] [PubMed]
- Nachon, F.; Carletti, E.; Ronco, C.; Trovaslet, M.; Nicolet, Y.; Jean, L.; Renard, P. Crystal structures of human cholinesterases in complex with huprine W and tacrine: Elements of specificity for anti-Alzheimer’s drugs targeting acetyl- and butyryl-cholinesterase. Biochem. J. 2013, 453, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Nicholls, A.; McGaughey, G.B.; Sheridan, R.P.; Good, A.C.; Warren, G.; Mathieu, M.; Muchmore, S.W.; Brown, S.P.; Grant, J.A.; Haigh, J.A.; et al. Molecular shape and medicinal chemistry: A perspective. J. Med. Chem. 2010, 53, 3862–3886. [Google Scholar] [CrossRef] [PubMed]
- Goodsell, D.S.; Morris, G.M.; Olson, A.J. Automated docking of flexible ligands: Applications of AutoDock. J. Mol. Recognit. 1996, 9, 1–5. [Google Scholar] [CrossRef]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [PubMed]
- Kryger, G.; Silman, I.; Sussman, J.L. Structure of acetylcholinesterase complexed with E2020 (Aricept®): Implications for the design of new anti-Alzheimer drugs. Structure 1999, 7, 297–307. [Google Scholar] [CrossRef]
- Wong, K.K.K.; Ngo, J.C.K.; Liu, S.; Lin, H.Q.; Hu, C.; Shaw, P.C.; Wan, D.C.C. Interaction study of two diterpenes, cryptotanshinone and dihydrotanshinone, to human acetylcholinesterase and butyrylcholinesterase by molecular docking and kinetic analysis. Chem. Biol. Interact. 2010, 187, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Youn, K.; Lee, J.; Ho, C.T.; Jun, M. Discovery of polymethoxyflavones from black ginger (Kaempferia parviflora) as potential β-secretase (BACE1) inhibitors. J. Funct. Foods 2016, 20, 567–574. [Google Scholar] [CrossRef]
Sample Availability: Samples of all the compounds used in this research are available from the authors and commercial sources. |






| Samples | EC50 (μM) a | |
|---|---|---|
| DPPH | Peroxynitrite | |
| Coumestrol | 53.98 ± 1.00 | 1.17 ± 0.11 |
| Puerarol | 82.55 ± 1.33 | 6.99 ± 0.30 |
| L-Ascorbic acid b | 17.37 ± 0.32 | ‒ |
| L-Penicillamine b | ‒ | 6.90 ± 0.08 |
| Samples | IC50 (µM) a | Inhibition Modes | Ki Values b | |||
|---|---|---|---|---|---|---|
| AChE | BChE | BACE1 | Kic | Kiu | ||
| Coumestrol | 42.33 ± 1.29 | 24.64 ± 2.28 | 51.04 ± 1.86 | Competitive c Mixed-type d | 48.91 12.07 | ‒ 41.87 |
| Puerarol | 144.80 ± 2.46 | >200 | 28.17 ± 2.48 | Mixed-type e | 33.8 | 73.19 |
| Berberine f | 2.22 ± 0.02 | 12.30 ± 1.15 | ‒ | ‒ | ‒ | ‒ |
| Quercetin f | ‒ | ‒ | 21.28 ± 1.42 | ‒ | ‒ | ‒ |
| Compounds | Binding Score (kcal/mol) | H-Bonds Interacting Residues (No. of H-bond) | Hydrophobic Interacting Residues |
|---|---|---|---|
| AChE (1ACJ) | |||
| Coumestrol | −8.63 | Glu199 (1) | Trp84, Gly117, Tyr130, Phe330, Tyr334, Trp432, Met436, Ile439, His440, Gly441, Tyr442 |
| Tacrine a (Catalytic inhibitor) | −9.80 | His440 (1) | Tyr442, Phe330, Trp84, Gly118, Trp432, Gly441, Tyr334, Glu199 |
| Donepezil a (Allosteric inhibitor) | −10.6 | ‒ | Tyr70, Ile275, Asp276, Trp279, Ile287, Phe288, Arg289, Tyr334, Tyr121, Ser286, Phe290, Phe330, Phe331 |
| BChE (4BDS) | |||
| Coumestrol (Catalytic inhibition mode) | −8.28 | Glu197 (1), Ser198 (1), Leu286 (1), His438 (1) | Trp82, Gly116, Gly117, Trp231, Phe398, Ile442 |
| Coumestrol (Allosteric inhibition mode) | −8.67 | Asp70 (1), Glu197 (1) | Gln67, Ile69, Trp82, Asn83, Gly115, Thr120, Gly121, Tyr128, Gly439, Try440 |
| Tacrine a (Catalytic inhibitor) | −8.60 | His438 (1) | Tyr332, Trp430, Trp82, Ala328, Glu197 |
| Cryptotanshinone a (Allosteric inhibitor) | −7.80 | ‒ | Asp70, Try82, Ala328, Tyr332, Trp430, Tyr440 |
| Compounds | Binding Score (kcal/mol) | H-Bonds Interacting Residues (No. of H-bond) | Hydrophobic Interacting Residues |
|---|---|---|---|
| Puerarol (Catalytic inhibition mode) | −8.80 | Thr231 (1) | Asp32, Gly34, Val69, Tyr71, Trp76, Phe108, Trp115, Ile118, Asp228 |
| Puerarol (Allosteric inhibition mode) | −8.03 | Thr232 (1) | Ser10, Gly156, Ala157, Trp277, Gln303, Gln304, Arg307, Pro308, Asp318, Tyr320, Ala335, Val336, Gln339, Val361, |
| QUD a (Catalytic inhibitor) | −9.30 | Asp228 (1), Asp32 (2), Gly230 (1) | Lys107, Lys75, Gly74, Leu30, Thr231, Val69, Tyr198, Ile226, Thr329, Gly34, Arg235, Ser35, Tyr71, Ile118 |
| TMF a,b (Allosteric inhibitor) | −7.80 | Gly11 (1) | Ser10, Tyr14, Thr232, Trp277, Glu303, Gln304, Leu306, Arg307, Pro308, Tyr320, Ala335, Val336, Glu339 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koirala, P.; Seong, S.H.; Jung, H.A.; Choi, J.S. Comparative Evaluation of the Antioxidant and Anti-Alzheimer’s Disease Potential of Coumestrol and Puerarol Isolated from Pueraria lobata Using Molecular Modeling Studies. Molecules 2018, 23, 785. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules23040785
Koirala P, Seong SH, Jung HA, Choi JS. Comparative Evaluation of the Antioxidant and Anti-Alzheimer’s Disease Potential of Coumestrol and Puerarol Isolated from Pueraria lobata Using Molecular Modeling Studies. Molecules. 2018; 23(4):785. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules23040785
Chicago/Turabian StyleKoirala, Prashamsa, Su Hui Seong, Hyun Ah Jung, and Jae Sue Choi. 2018. "Comparative Evaluation of the Antioxidant and Anti-Alzheimer’s Disease Potential of Coumestrol and Puerarol Isolated from Pueraria lobata Using Molecular Modeling Studies" Molecules 23, no. 4: 785. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules23040785





