In Vitro and In Silico Studies of the Molecular Interactions of Epigallocatechin-3-O-gallate (EGCG) with Proteins That Explain the Health Benefits of Green Tea
Abstract
:1. Introduction
2. Affinity Gel Chromatography (AGC) and Other Conventional Methods
2.1. Studies on the Interaction between Catechins and Blood Proteins
2.2. Interaction between Catechins and Cancer-Related Protein
2.3. Interaction between Catechins and Proteins Related to Cardiac Muscle Disease and Amyloid Disease
3. Surface Plasmon Resonance (SPR)
3.1. Interaction between Catechins and Proteins Related to Cancer, Inflammatory Disease, and Oral Health
3.2. Interaction between Catechins and Proteins Related to Inflammatory Disease and Oral Health
4. Computational MDA
4.1. Interaction between Catechins and Cancer-Related Proteins
4.2. Interaction between Catechins and MetS-Related Proteins
4.3. Interaction between Catechins and Inflammation-Related Proteins
4.4. Interaction between Catechins and Microbial Proteins
4.5. Interaction between Catechins and Proteins Related to Neurodegenerative Diseases
5. X-ray Crystallographic Analysis of the Catechin–Protein Complex
5.1. Interaction between Catechins and Cancer-Related Proteins
5.2. Interaction between Catechins and Aamyloidosis Protein
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 11bHD1 | 11β-hydroxysteroid dehydrogenase type 1 |
| 67LR | 67-kDa laminin receptor |
| Aβ | amyloid β protein |
| ACE | angiotensin converting enzyme |
| AD | Alzheimer’s disease |
| AGC | affinity gel chromatography |
| AhR | aryl hydrocarbon receptor |
| AMPK | AMP-activated protein kinase |
| aSMase | acid sphingomyelinase |
| Bcl2 | B-cell lymphoma-2 |
| BclxL | B-cell lymphoma-extra large |
| BSA | bovine serum albumin |
| CT | cholera toxin |
| CXCL | C-X-C motif chemokine ligand |
| DENV | Dengue virus |
| DHFR | dihydrofolate reductase |
| DNMT | DNA methyltranseferase |
| EC | (−)-epicatechin |
| ECG | (−)-epicatechin gallate |
| EGC | (−)-epigallocatechin |
| EGCG | (−)-epigallocatechin-3-O-gallate |
| EGE | envelope glycoprotein E |
| EGF | epidermal growth factor |
| FASN | fatty acid synthase |
| G3BP1 | SH3 domain-binding protein 1 |
| GAP | GTPase-activating protein |
| GBM | glioblastoma |
| GDH | glutamate dehydrogenase |
| GLIDE | grid-based ligand docking with energetics |
| GRP78 | glucose-regulated protein 78 kDa |
| GST | glutathione-S-transferase |
| GTCs | green tea catechins |
| HDAC | histone deacetylase |
| HFD | high fat diet |
| HHS | hyperinsulinism/hyperammonemia syndrome |
| HMGR | hydroxymethyl-glutaryl-CoA reductase |
| hMLH1 | human mutL homologue 1 |
| HPV | human papillomavirus |
| HSP | heat shock protein |
| IGF1R | insulin-like growth factor 1 receptor |
| IL | interleukin |
| MAPK | mitogen-activated protein kinase |
| MDA | molecular docking analysis |
| MetS | metabolic syndrome |
| MMP | matrix metalloproteinase |
| MT1-MMP | membrane-type 1 matrix metalloproteinase |
| NF-κB | nuclear factor-kappa B |
| PGG | penta-O-galloyl-β-d-glucose |
| PI3K | phosphoinositide-3-kinase |
| Pin1 | peptidyl prolyl cis/trans isomerase 1 |
| PP1 | protein phosphatase-1 |
| PP2A | protein phosphatase-2A |
| QCM | quartz crystal microbalance |
| RARβ | retinoic acid receptor beta |
| ROS | reactive oxygen species |
| SH3 | Src homology 3 |
| SPR | surface plasmon resonance |
| SREBP | sterol-response element binding protein |
| STAT | signal transducer and activator of transcription |
| TAK1 | transforming growth factor β-activated kinase 1 |
| TGFRII | transforming growth factor β type II receptor |
| TGFβ | transforming growth factor β |
| TNF | tumor necrosis factor |
| TRAF6 | TNF receptor associated factor 6 |
| TTR | transthyretin |
| Ubc13 | ubiquitin-conjugating protein 13 |
| VEGF | vascular endothelial growth factor |
| VEGF1R | vascular endothelial growth factor 1 receptor |
| VEGFR2 | VEGF-induced VEGF receptor-2 |
| ZAP-70 | ζ chain-associated 70 kDa protein |
References
- Hayakawa, S.; Saito, K.; Miyoshi, N.; Ohishi, T.; Oishi, Y.; Miyoshi, M.; Nakamura, Y. Anti-cancer effects of green tea by either anti- or pro- oxidative mechanisms. Asian Pac. J. Cancer Prev. 2016, 17, 1649–1654. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Pervin, M.; Goto, S.; Isemura, M.; Nakamura, Y. Beneficial effects of tea and the green tea catechin epigallocatechin-3-gallate on obesity. Molecules 2016, 21, 1305. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Miyoshi, N.; Isemura, M. Health-promoting effects of green tea. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2012, 88, 88–101. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.S.; Zhang, J.; Zhang, L.; Huang, J.; Wang, Y. Mechanisms of body weight reduction and metabolic syndrome alleviation by tea. Mol. Nutr. Food Res. 2016, 60, 160–174. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.S.; Wang, H. Cancer preventive activities of tea catechins. Molecules 2016, 21, 1679. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.S.; Wang, H. Tea catechins in cancer prevention and therapy-Molecular mechanism and human relevance. In Health Benefits of Green Tea: An Evidence-Based Approach; Hara, Y., Yang, C.S., Isemura, M., Tomita, I., Eds.; Oxfordshire: Oxford, UK, 2016; pp. 65–83. [Google Scholar]
- Suzuki, T.; Miyoshi, N.; Hayakawa, S.; Imai, S.; Isemura, M.; Nakamura, Y. Health Benefits of Tea Consumption. In Beverage Impacts on Health and Nutrition; Wilson, T., Templ, N.J., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 29–47. [Google Scholar]
- Bettuzzi, S.; Brausi, M.; Rizzi, F.; Castagnetti, G.; Peracchia, G.; Corti, A. Chemoprevention of human prostate cancer by oral administration of green tea catechins in volunteers with high-grade prostate intraepithelial neoplasia: A preliminary report from a one-year proof-of-principle study. Cancer Res. 2006, 66, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- Hara, Y. Tea catechins and their applications as supplements and pharmaceutics. Pharmacol. Res. 2011, 64, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Nagao, T.; Hase, T.; Tokimitsu, I. A green tea extract high in catechins reduces body fat and cardiovascular risks in humans. Obesity 2007, 15, 1473–1483. [Google Scholar] [CrossRef] [PubMed]
- Scholey, A.; Downey, L.A.; Ciorciari, J.; Pipingas, A.; Nolidin, K.; Finn, M.; Wines, M.; Catchlove, S.; Terrens, A.; Barlow, E.; et al. Acute neurocognitive effects of epigallocatechin gallate (EGCG). Appetite 2012, 58, 767–770. [Google Scholar] [CrossRef] [PubMed]
- Fujiki, H.; Suganuma, M.; Okabe, S.; Sueoka, N.; Komori, A.; Sueoka, E.; Kozu, T.; Tada, Y.; Suga, K.; Imai, K.; et al. Cancer inhibition by green tea. Mutat. Res. 1998, 402, 307–310. [Google Scholar] [CrossRef]
- Maeda-Yamamoto, M.; Kawahara, H.; Tahara, N.; Tsuji, K.; Hara, Y.; Isemura, M. Effects of tea polyphenols on the invasion and matrix metalloproteinases activities of human fibrosarcoma HT1080 cells. J. Agric. Food Chem. 1999, 47, 2350–2354. [Google Scholar] [CrossRef] [PubMed]
- Hastak, K.; Gupta, S.; Ahmad, N.; Agarwal, M.K.; Agarwal, M.L.; Mukhtar, H. Role of p53 and NF-κB in epigallocatechin-3-gallate-induced apoptosis of LNCaP cells. Oncogene 2003, 22, 4851–4859. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Bao, S.; Yang, W.; Zhang, J.; Li, L.; Shan, Z.; Teng, W. Epigallocatechin gallate prevents inflammation by reducing macrophage infiltration and inhibiting tumor necrosis factor-α signaling in the pancreas of rats on a high-fat diet. Nutr. Res. 2014, 34, 1066–1074. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.X. Inhibition of fatty acid synthase by polyphenols. Curr. Med. Chem. 2006, 13, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.B.; Chern, T.R.; Wei, T.T.; Chen, C.C.; Lin, J.H.; Fang, J.M. Design and synthesis of dual-action inhibitors targeting histone deacetylases and 3-hydroxy-3-methylglutaryl coenzyme A reductase for cancer treatment. J. Med. Chem. 2013, 56, 3645–3655. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Dey, S.K.; Saha, C. Inhibition of catalase by tea catechins in free and cellular state: A biophysical approach. PLoS ONE 2014, 9, e102460. [Google Scholar] [CrossRef] [PubMed]
- Arora, J.P.; Singhal, V.K.; Chand, M.; Laxmi; Pal, C. Study of interaction between catechin and native and modified bovine serum albumin by physico-chemical methods. Indian J. Biochem. Biophys. 1989, 26, 14–18. [Google Scholar] [PubMed]
- Minoda, K.; Ichikawa, T.; Katsumata, T.; Onobori, K.; Mori, T.; Suzuki, Y.; Ishii, T.; Nakayama, T. Influence of the galloyl moiety in tea catechins on binding affinity for human serum albumin. J. Nutr. Sci. Vitaminol. 2010, 56, 331–334. [Google Scholar] [CrossRef] [PubMed]
- Sazuka, M.; Itoi, T.; Suzuki, Y.; Odani, S.; Koide, T.; Isemura, M. Evidence for the interaction between (−)-epigallocatechin gallate and human plasma proteins fibronectin, fibrinogen, and histidine-rich glycoprotein. Biosci. Biotechnol. Biochem. 1996, 60, 1317–1319. [Google Scholar] [CrossRef] [PubMed]
- Sazuka, M.; Isemura, M.; Isemura, S. Interaction between the carboxyl-terminal heparin-binding domain of fibronectin and (−)-epigallocatechin gallate. Biosci. Biotechnol. Biochem. 1998, 62, 1031–1032. [Google Scholar] [CrossRef] [PubMed]
- Palermo, C.M.; Westlake, C.A.; Gasiewicz, T.A. Epigallocatechin gallate inhibits aryl hydrocarbon receptor gene transcription through an indirect mechanism involving binding to a 90 kDa heat shock protein. Biochemistry 2005, 44, 5041–5052. [Google Scholar] [CrossRef] [PubMed]
- Moses, M.A.; Henry, E.C.; Ricke, W.A.; Gasiewicz, T.A. The heat shock protein 90 inhibitor, (−)-epigallocatechin gallate, has anticancer activity in a novel human prostate cancer progression model. Cancer Prev. Res. 2015, 8, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Ermakova, S.P.; Kang, B.S.; Choi, B.Y.; Choi, H.S.; Schuster, T.F.; Ma, W.Y.; Bode, A.M.; Dong, Z. (−)-Epigallocatechin gallate overcomes resistance to etoposide-induced cell death by targeting the molecular chaperone glucose-regulated protein 78. Cancer Res. 2006, 66, 9260–9269. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; He, Z.; Ermakova, S.; Zheng, D.; Tang, F.; Cho, Y.Y.; Zhu, F.; Ma, W.Y.; Sham, Y.; Rogozin, E.A.; et al. Direct inhibition of insulin-like growth factor-I receptor kinase activity by (−)-epigallocatechin-3-gallate regulates cell transformation. Cancer Epidemiol. Biomarkers Prev. 2007, 16, 598–605. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Tang, F.; Ermakova, S.; Li, M.; Zhao, Q.; Cho, Y.Y.; Ma, W.Y.; Choi, H.S.; Bode, A.M.; Yang, C.S.; et al. Fyn is a novel target of (−)-epigallocatechin gallate in the inhibition of JB6 Cl41 cell transformation. Mol. Carcinog. 2008, 47, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.H.; Choi, H.S.; Pugliese, A.; Lee, S.Y.; Chae, J.I.; Choi, B.Y.; Bode, A.M.; Dong, Z. (−)-Epigallocatechin gallate regulates CD3-mediated T cell receptor signaling in leukemia through the inhibition of ZAP-70 kinase. J. Biol. Chem. 2008, 283, 28370–28379. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.H.; Su, Z.Y.; Chae, J.I.; Kim, D.J.; Zhu, F.; Ma, W.Y.; Bode, A.M.; Yang, C.S.; Dong, Z. Epigallocatechin gallate suppresses lung cancer cell growth through Ras-GTPase-activating protein SH3 domain-binding protein 1. Cancer Prev. Res. 2010, 3, 670–679. [Google Scholar] [CrossRef] [PubMed]
- Urusova, D.V.; Shim, J.H.; Kim, D.J.; Jung, S.K.; Zykova, T.A.; Carper, A.; Bode, A.M.; Dong, Z. Epigallocatechin-gallate suppresses tumorigenesis by directly targeting Pin1. Cancer Prev. Res. 2011, 4, 1366–1377. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lei, Z.; Huang, Z.; Zhang, X.; Zhou, Y.; Luo, Z.; Zeng, W.; Su, J.; Peng, C.; Chen, X. Epigallocatechin-3-gallate(EGCG) suppresses melanoma cell growth and metastasis by targeting TRAF6 activity. Oncotarget 2016, 7, 79557–79571. [Google Scholar] [CrossRef] [PubMed]
- Sazuka, M.; Imazawa, H.; Shoji, Y.; Mita, T.; Hara, Y.; Isemura, M. Inhibition of collagenases from mouse lung carcinoma cells by green tea catechins and black tea theaflavins. Biosci. Biotechnol. Biochem. 1997, 61, 1504–1506. [Google Scholar] [CrossRef] [PubMed]
- Ermakova, S.; Choi, B.Y.; Choi, H.S.; Kang, B.S.; Bode, A.M.; Dong, Z. The intermediate filament protein vimentin is a new target for epigallocatechin gallate. J. Biol. Chem. 2005, 280, 16882–16890. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, S.; Saeki, K.; Sazuka, M.; Suzuki, Y.; Shoji, Y.; Ohta, T.; Kaji, K.; Yuo, A.; Isemura, M. Apoptosis induction by epigallocatechin gallate involves its binding to Fas. Biochem. Biophys. Res. Commun. 2001, 285, 1102–1106. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Isemura, M. Inhibitory effect of epigallocatechin gallate on adhesion of murine melanoma cells to laminin. Cancer Lett. 2001, 173, 15–20. [Google Scholar] [CrossRef]
- Suzuki, Y.; Isemura, M. Binding interaction between (−)-epigallocatechin gallate causes impaired spreading of cancer cells on fibrinogen. Biomed. Res. 2013, 34, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Suzuki, T.; Minami, T.; Isemura, M. Involvement of impaired interaction with β1 integrin in epigallocatechin gallate-mediated inhibition of fibrosarcoma HT-1080 cell adhesion to fibronectin. J. Health Sci. 2006, 52, 103–109. [Google Scholar] [CrossRef]
- Suzuki, Y.; Hattori, S.; Isemura, M. Epigallocatechin-3-O-gallate inhibits fibroblast contraction of floating collagen gel: Interaction between epigallocatechin-3-O-gallate and platelet derived growth factor. Biosci. Biotechnol. Biochem. 2004, 68, 1817–1820. [Google Scholar] [CrossRef] [PubMed]
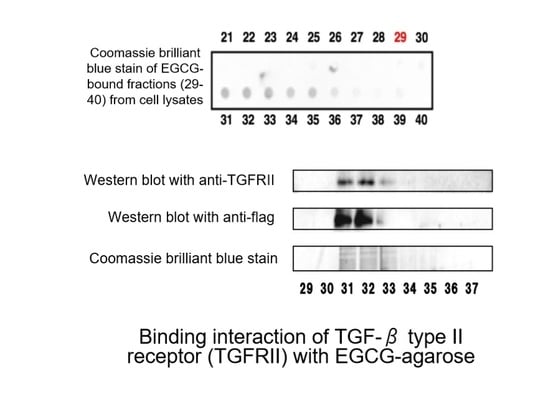
- Tabuchi, M.; Hayakawa, S.; Honda, E.; Ooshima, K.; Itoh, T.; Yoshida, K.; Park, A.-M.; Higashino, H.; Isemura, M.; Munakata, H. Epigallocatechin-3-gallate suppresses transforming growth factor-β signaling by interacting with the transforming growth factor-β type II receptor. World J. Exp. Med. 2013, 3, 100. [Google Scholar] [CrossRef]
- Barrack, E.R. TGF beta in prostate cancer: A growth inhibitor that can enhance tumorigenicity. Prostate 1997, 31, 61–70. [Google Scholar] [CrossRef]
- Rodon, J.; Carducci, M.A.; Sepulveda-Sanchez, J.M.; Azaro, A.; Calvo, E.; Seoane, J.; Brana, I.; Sicart, E.; Gueorguieva, I.; Cleverly, A.L.; et al. First-in-human dose study of the novel transforming growth factor-β receptor I kinase inhibitor LY2157299 monohydrate in patients with advanced cancer and glioma. Clin. Cancer Res. 2015, 21, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Ishii, T.; Mizuno, D.; Mori, T.; Yamaji, R.; Nakamura, Y.; Kumazawa, S.; Nakayama, T.; Akagawa, M. (−)-Epigallocatechin-3-gallate suppresses growth of AZ521 human gastric cancer cells by targeting the DEAD-box RNA helicase p68. Free Radic. Biol. Med. 2011, 50, 1324–1335. [Google Scholar] [CrossRef] [PubMed]
- Tadano, N.; Du, C.K.; Yumoto, F.; Morimoto, S.; Ohta, M.; Xie, M.F.; Nagata, K.; Zhan, D.Y.; Lu, Q.W.; Miwa, Y.; et al. Biological actions of green tea catechins on cardiac troponin C. Br. J. Pharmacol. 2010, 161, 1034–1043. [Google Scholar] [CrossRef] [PubMed]
- Kamihira-Ishijima, M.; Nakazawa, H.; Kira, A.; Naito, A.; Nakayama, T. Inhibitory mechanism of pancreatic amyloid fibril formation: Formation of the complex between tea catechins and the fragment of residues 22–27. Biochemistry. 2012, 51, 10167–10174. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, H.; Koga, K.; Fujimura, Y.; Yamada, K. A receptor for green tea polyphenol EGCG. Nat. Struct. Mol. Biol. 2004, 11, 380–381. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, H. Green tea polyphnol EGCG-sensing Receptor. In Health Benefits of Green Tea: An Evidence-Based Approach; Hara, Y., Yang, C.S., Isemura, M., Tomita, I., Eds.; Oxfordshire: Oxford, UK, 2016; pp. 89–100. [Google Scholar]
- Fujimura, Y.; Sumida, M.; Sugihara, K.; Tsukamoto, S.; Yamada, K.; Tachibana, H. Green tea polyphenol EGCG sensing motif on the 67-kDa laminin receptor. PLoS ONE 2012, 7, e37942. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, S.; Hirotsu, K.; Kumazoe, M.; Goto, Y.; Sugihara, K.; Suda, T.; Tsurudome, Y.; Suzuki, T.; Yamashita, S.; Kim, Y.; et al. Green tea polyphenol EGCG induces lipid-raft clustering and apoptotic cell death by activating protein kinase Cδ and acid sphingomyelinase through a 67 kDa laminin receptor in multiple myeloma cells. Biochem. J. 2012, 443, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, H. Green tea polyphenol sensing. Proc. Jpn. Acad. Ser. B 2011, 87, 66–80. [Google Scholar] [CrossRef]
- Trnkova, L.; Ricci, D.; Grillo, C.; Colotti, G.; Altieri, F. Green tea catechins can bind and modify ERp57/PDIA3 activity. Biochim. Biophys. Acta 2013, 1830, 2671–2682. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Alcorn, J.F.; Craigo, J.K.; Tjoeng, C.; Tarwater, P.M.; Kolls, J.K.; Reinhart, T.A. Epigallocatechin-3-gallate reduces airway inflammation in mice through binding to proinflammatory chemokines and inhibiting inflammatory cell recruitment. J. Immunol. 2011, 186, 3693–3700. [Google Scholar] [CrossRef] [PubMed]
- Hara, K.; Ohara, M.; Hayashi, I.; Hino, T.; Nishimura, R.; Iwasaki, Y.; Ogawa, T.; Ohyama, Y.; Sugiyama, M.; Amano, H. The green tea polyphenol (−)-epigallocatechin gallate precipitates salivary proteins including alpha-amylase: Biochemical implications for oral health. Eur. J. Oral. Sci. 2012, 120, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, A.; Nandy, S.K.; Sarkar, J.; Chakraborti, T.; Chakraborti, S. Inhibition of pro-/active MMP-2 by green tea catechins and prediction of their interaction by molecular docking studies. Mol. Cell Biochem. 2017, 427, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, J.; Nandy, S.K.; Chowdhury, A.; Chakraborti, T.; Chakraborti, S. Inhibition of MMP-9 by green tea catechins and prediction of their interaction by molecular docking analysis. Biomed. Pharmacother. 2016, 84, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Leone, M.; Zhai, D.; Sareth, S.; Kitada, S.; Reed, J.C.; Pellecchia, M. Cancer prevention by tea polyphenols is linked to their direct inhibition of antiapoptotic Bcl-2-family proteins. Cancer Res. 2003, 63, 8118–8121. [Google Scholar] [PubMed]
- Bhattacharjee, R.; Devi, A.; Mishra, S. Molecular docking and molecular dynamics studies reveal structural basis of inhibition and selectivity of inhibitors EGCG and OSU-03012 toward glucose regulated protein-78 (GRP78) overexpressed in glioblastoma. J. Mol. Model. 2015, 21, 272. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lee, A.S. Stress induction of GRP78/BiP and its role in cancer. Curr. Mol. Med. 2006, 6, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Bast, F. Screening of multi-targeted natural compounds for receptor tyrosine kinases inhibitors and biological evaluation on cancer cell lines, in silico and in vitro. Med. Oncol. 2015, 32, 233. [Google Scholar] [CrossRef] [PubMed]
- Hsu, K.C.; Sung, T.Y.; Lin, C.T.; Chiu, Y.Y.; Hsu, J.T.; Hung, H.C.; Sun, C.M.; Barve, I.; Chen, W.L.; Huang, W.C.; et al. Anchor-based classification and type-C inhibitors for tyrosine kinases. Sci. Rep. 2015, 5, 10938. [Google Scholar] [CrossRef] [PubMed]
- Jankun, J.; Selman, S.H.; Swiercz, R.; Skrzypczak-Jankun, E. Why drinking green tea could prevent cancer. Nature 1997, 387, 561. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Yang, K.; Li, Y. Investigate the binding of catechins to trypsin using docking and molecular dynamics simulation. PLoS ONE 2015, 10, e0125848. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.M.; Daniel, K.G.; Wang, Z.; Guida, W.C.; Chan, T.H.; Dou, Q.P. Docking studies and model development of tea polyphenol proteasome inhibitors: Applications to rational drug design. Proteins 2004, 54, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Chen, T.S.; Lu, T. A comparative reverse docking strategy to identify potential antineoplastic targets of tea functional components and binding mode. Int. J. Mol. Sci. 2011, 12, 5200–5212. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.Z.; Wang, Y.; Ai, N.; Hou, Z.; Sun, Y.; Lu, H.; Welsh, W.; Yang, C.S. Tea polyphenol (−)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003, 63, 7563–7570. [Google Scholar] [PubMed]
- Khan, M.A.; Hussain, A.; Sundaram, M.K.; Alalami, U.; Gunasekera, D.; Ramesh, L.; Hamza, A.; Quraishi, U. (−)-Epigallocatechin-3-gallate reverses the expression of various tumor-suppressor genes by inhibiting DNA methyltransferases and histone deacetylases in human cervical cancer cells. Oncol. Rep. 2015, 33, 1976–1984. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-del-Campo, L.; Saez-Ayala, M.; Chazarra, S.; Cabezas-Herrera, J.; Rodriguez-Lopez, J.N. Binding of natural and synthetic polyphenols to human dihydrofolate reductase. Int. J. Mol. Sci. 2009, 10, 5398–5410. [Google Scholar] [CrossRef] [PubMed]
- Van Aller, G.S.; Carson, J.D.; Tang, W.; Peng, H.; Zhao, L.; Copeland, R.A.; Tummino, P.J.; Luo, L. Epigallocatechin gallate (EGCG), a major component of green tea, is a dual phosphoinositide-3-kinase/mTOR inhibitor. Biochem. Biophys. Res. Commun. 2011, 406, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Kiss, A.; Becsi, B.; Kolozsvari, B.; Komaromi, I.; Kover, K.E.; Erdodi, F. Epigallocatechin-3-gallate and penta-O-galloyl-β-d-glucose inhibit protein phosphatase-1. FEBS J. 2013, 280, 612–626. [Google Scholar] [CrossRef] [PubMed]
- Moyle, C.W.; Cerezo, A.B.; Winterbone, M.S.; Hollands, W.J.; Alexeev, Y.; Needs, P.W.; Kroon, P.A. Potent inhibition of VEGFR-2 activation by tight binding of green tea epigallocatechin gallate and apple procyanidins to VEGF: Relevance to angiogenesis. Mol. Nutr. Food Res. 2015, 59, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ren, X.; Deng, C.; Yang, L.; Yan, E.; Guo, T.; Li, Y.; Xu, M.X. Mechanism of the inhibition of the STAT3 signaling pathway by EGCG. Oncol. Rep. 2013, 30, 2691–2696. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; He, W.; Yao, L.; Zhang, H.; Liu, Z.; Wang, W.; Ye, Y.; Cao, J. Characterization of binding interactions of (−)-epigallocatechin-3-gallate from green tea and lipase. J. Agric. Food Chem. 2013, 61, 8829–8835. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Sun, Z.; Dong, S.; Liu, Y.; Liu, Y. Molecular interactions between (−)-epigallocatechin gallate analogs and pancreatic lipase. PLoS ONE 2014, 9, e111143. [Google Scholar] [CrossRef] [PubMed]
- Cuccioloni, M.; Mozzicafreddo, M.; Spina, M.; Tran, C.N.; Falconi, M.; Eleuteri, A.M.; Angeletti, M. Epigallocatechin-3-gallate potently inhibits the in vitro activity of hydroxy-3-methyl-glutaryl-CoA reductase. J. Lipid Res. 2011, 52, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Islam, B.; Sharma, C.; Adem, A.; Aburawi, E.; Ojha, S. Insight into the mechanism of polyphenols on the activity of HMGR by molecular docking. Drug Des. Devel. Ther. 2015, 9, 4943–4951. [Google Scholar] [PubMed]
- Hintzpeter, J.; Stapelfeld, C.; Loerz, C.; Martin, H.J.; Maser, E. Green tea and one of its constituents, Epigallocatechine-3-gallate, are potent inhibitors of human 11β-hydroxysteroid dehydrogenase type 1. PLoS ONE 2014, 9, e84468. [Google Scholar] [CrossRef] [PubMed]
- Ke, Z.; Su, Z.; Zhang, X.; Cao, Z.; Ding, Y.; Cao, L.; Ding, G.; Wang, Z.; Liu, H.; Xiao, W. Discovery of a potent angiotensin converting enzyme inhibitor via virtual screening. Bioorg. Med. Chem. Lett. 2017, 27, 3688–3692. [Google Scholar] [CrossRef] [PubMed]
- Menegazzi, M.; Mariotto, S.; Dal Bosco, M.; Darra, E.; Vaiana, N.; Shoji, K.; Safwat, A.A.; Marechal, J.D.; Perahia, D.; Suzuki, H.; Romeo, S. Direct interaction of natural and synthetic catechins with signal transducer activator of transcription 1 affects both its phosphorylation and activity. FEBS J. 2014, 281, 724–738. [Google Scholar] [CrossRef] [PubMed]
- Townsend, P.A.; Scarabelli, T.M.; Pasini, E.; Gitti, G.; Menegazzi, M.; Suzuki, H.; Knight, R.A.; Latchman, D.S.; Stephanou, A. Epigallocatechin-3-gallate inhibits STAT-1 activation and protects cardiac myocytes from ischemia/reperfusion-induced apoptosis. FASEB J. 2004, 18, 1621–1623. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Umar, S.; Riegsecker, S.; Chourasia, M.; Ahmed, S. Regulation of transforming growth factor beta-activated kinase activation by epigallocatechin-3-gallate in rheumatoid arthritis synovial fibroblasts: Suppression of K(63)-linked autoubiquitination of tumor necrosis factor receptor-associated factor 6. Arthritis Rheumatol. 2016, 68, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Fechtner, S.; Singh, A.; Chourasia, M.; Ahmed, S. Molecular insights into the differences in anti-inflammatory activities of green tea catechins on IL-1β signaling in rheumatoid arthritis synovial fibroblasts. Toxicol. Appl. Pharmacol. 2017, 329, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Toda, M.; Okubo, S.; Ikigai, H.; Suzuki, T.; Suzuki, Y.; Hara, Y.; Shimamura, T. The protective activity of tea catechins against experimental infection by Vibrio cholerae O1. Microbiol. Immunol. 1992, 36, 999–1001. [Google Scholar] [CrossRef] [PubMed]
- Cherubin, P.; Garcia, M.C.; Curtis, D.; Britt, C.B.; Craft, J.W., Jr.; Burress, H.; Berndt, C.; Reddy, S.; Guyette, J.; Zheng, T.; et al. Inhibition of cholera toxin and other AB toxins by polyphenolic compounds. PLoS ONE 2016, 11, e0166477. [Google Scholar] [CrossRef] [PubMed]
- Ismail, N.A.; Jusoh, S.A. Molecular docking and molecular dynamics simulation studies to predict flavonoid binding on the surface of DENV2 E protein. Interdiscip. Sci. 2017, 9, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Jena, L.; Galande, S.; Daf, S.; Mohod, K.; Varma, A.K. Elucidating molecular interactions of natural inhibitors with HPV-16 E6 oncoprotein through docking analysis. Genomics Inform. 2014, 12, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Khalid, S.; Paul, S. Identifying a C-terminal ATP binding sites-based novel Hsp90-Inhibitor in silico: A plausible therapeutic approach in Alzheimer’s disease. Med. Hypotheses. 2014, 83, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Ehrnhoefer, D.E.; Bieschke, J.; Boeddrich, A.; Herbst, M.; Masino, L.; Lurz, R.; Engemann, S.; Pastore, A.; Wanker, E.E. EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat. Struct. Mol. Biol. 2008, 15, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Chebaro, Y.; Jiang, P.; Zang, T.; Mu, Y.; Nguyen, P.H.; Mousseau, N.; Derreumaux, P. Structures of Aβ17–42 trimers in isolation and with five small-molecule drugs using a hierarchical computational procedure. J. Phys. Chem. B 2012, 116, 8412–8422. [Google Scholar] [CrossRef] [PubMed]
- Lemkul, J.A.; Bevan, D.R. The role of molecular simulations in the development of inhibitors of amyloid β-peptide aggregation for the treatment of Alzheimer’s disease. ACS Chem. Neurosci. 2012, 3, 845–856. [Google Scholar] [CrossRef] [PubMed]
- Hyung, S.J.; DeToma, A.S.; Brender, J.R.; Lee, S.; Vivekanandan, S.; Kochi, A.; Choi, J.S.; Ramamoorthy, A.; Ruotolo, B.T.; Lim, M.H. Insights into antiamyloidogenic properties of the green tea extract (−)-epigallocatechin-3-gallate toward metal-associated amyloid-β species. Proc. Nat. Acad. Sci. USA 2013, 110, 3743–3748. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, M.; Chen, P.; Narayan, S.; Matschinsky, F.M.; Bennett, M.J.; Stanley, C.A.; Smith, T.J. Green tea polyphenols control dysregulated glutamate dehydrogenase in transgenic mice by hijacking the ADP activation site. J. Biol. Chem. 2011, 286, 34164–34174. [Google Scholar] [CrossRef] [PubMed]
- Miyata, M.; Sato, T.; Kugimiya, M.; Sho, M.; Nakamura, T.; Ikemizu, S.; Chirifu, M.; Mizuguchi, M.; Nabeshima, Y.; Suwa, Y.; et al. The crystal structure of the green tea polyphenol (−)-epigallocatechin gallate-transthyretin complex reveals a novel binding site distinct from the thyroxine binding site. Biochemistry 2010, 49, 6104–6114. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]




| Experimental Mode | Protein | Binding Characteristics | Head Author (Year) | Reference |
|---|---|---|---|---|
| AGC | Fibronectin | EGCG binds to the carboxyl-terminal heparin-binding domain. | Sazuka, M. (1996;1998) | [21] [22] |
| AGC | MMP-2 * | EGCG binding to MMP-2 was identified by gelatin zymography. | Sazuka, M. (1997) | [32] |
| AGC | MMP-9 ** | EGCG binding to MMP-9 was identified by gelatin zymography. | Sazuka, M. (1997) | [32] |
| AGC, PD | Vimentin | EGCG binds to the region of 50–63 residues. | Ermakova, S. (2005) | [33] |
| PD | HSP90 ** | EGCG binds to a C-terminal geldanamycin binding site (amino acid residues 538–728) | Palermo, C.M. (2005) Moses, M.A. (2015) | [23] [24] |
| PD | GRP78 ** | EGCG binds to the ATPase catalytic domain (211–654 residues) | Ermakova, S.P. (2006) | [25] |
| PD | IGF1R ** | The participating residues in the binding include Gln977, Lys1003, MEet1052, The1053, and Asp1123EGCG binds to the ATP binding pocket in β-subunit. | Li, M. (2007) | [26] |
| PD | Fyn | EGCG binds to the SH2 domain, but not the SH3 domain | He, Z. (2008) | [27] |
| PD | ZAP70 ** | EGCG binds to an ATP binding siteGlu415, Ala417, Lys369, Asp479, Glu386. | Shim, J.H. (2008) | [28] |
| PD | G3BP1 | EGCG binds to the region of amino acid residues 226–340. | Shim, J.H. (2010) | [29] |
| PD | Pin1 *** | EGCG bound to WW domain with two conserved tryptophans (1–39) pSer/Thr–Pro recognition loop of Met15–S16-R17-S18-R21-Tyr23 and to the peptidyl prolyl isomerase domain of Pin. EGCG creates several strong contacts with Pin1 at Asp112, Ser114, Trp73, and Ser114. | Urusova, D.V. (2011) | [30] |
| PD | TRAF6 ** | EGCG binds to TRAF6 at the residues of Gln54, Gly55, Asp57 ILe72, Cys73 and Lys96. Mutation of Gln54, Asp57, ILe72 in TRAF6 destroys EGCG binding to TRAF6. | Zhang, J. (2016) | [31] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saeki, K.; Hayakawa, S.; Nakano, S.; Ito, S.; Oishi, Y.; Suzuki, Y.; Isemura, M. In Vitro and In Silico Studies of the Molecular Interactions of Epigallocatechin-3-O-gallate (EGCG) with Proteins That Explain the Health Benefits of Green Tea. Molecules 2018, 23, 1295. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules23061295
Saeki K, Hayakawa S, Nakano S, Ito S, Oishi Y, Suzuki Y, Isemura M. In Vitro and In Silico Studies of the Molecular Interactions of Epigallocatechin-3-O-gallate (EGCG) with Proteins That Explain the Health Benefits of Green Tea. Molecules. 2018; 23(6):1295. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules23061295
Chicago/Turabian StyleSaeki, Koichi, Sumio Hayakawa, Shogo Nakano, Sohei Ito, Yumiko Oishi, Yasuo Suzuki, and Mamoru Isemura. 2018. "In Vitro and In Silico Studies of the Molecular Interactions of Epigallocatechin-3-O-gallate (EGCG) with Proteins That Explain the Health Benefits of Green Tea" Molecules 23, no. 6: 1295. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules23061295