Synthesis, Antimicrobial Activity and Molecular Docking of Novel Thiourea Derivatives Tagged with Thiadiazole, Imidazole and Triazine Moieties as Potential DNA Gyrase and Topoisomerase IV Inhibitors
Abstract
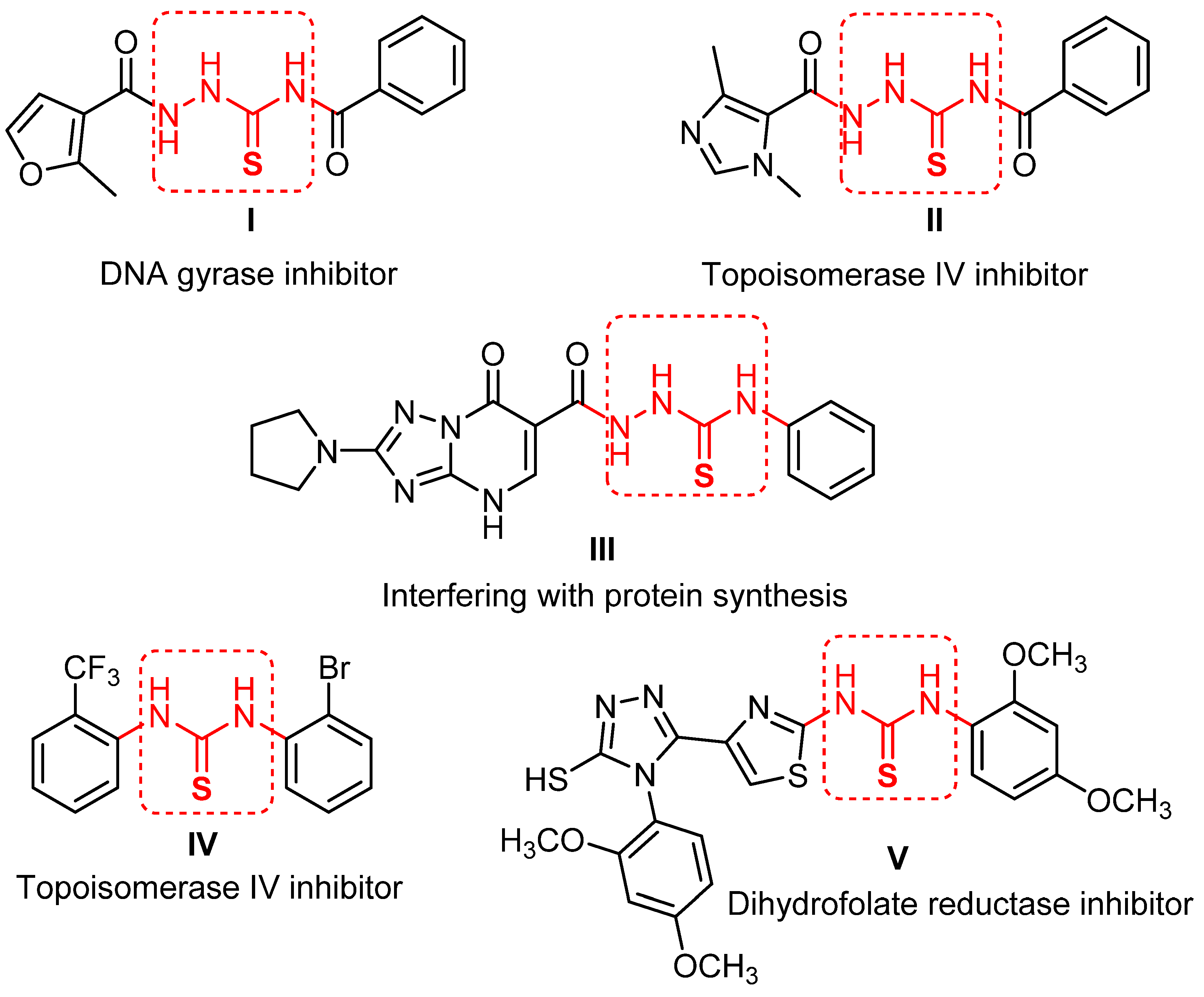
:1. Introduction
2. Results and Discussion
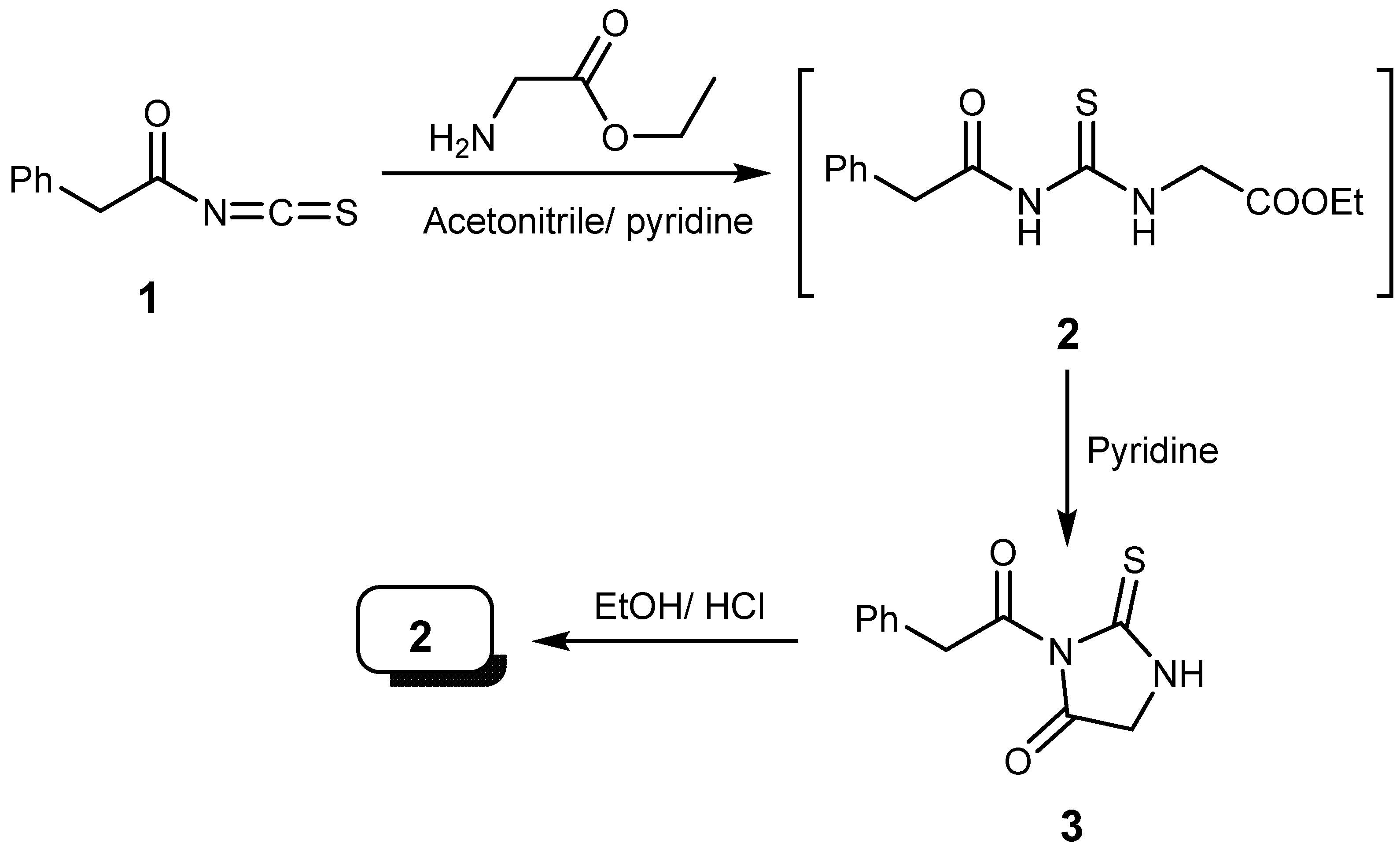
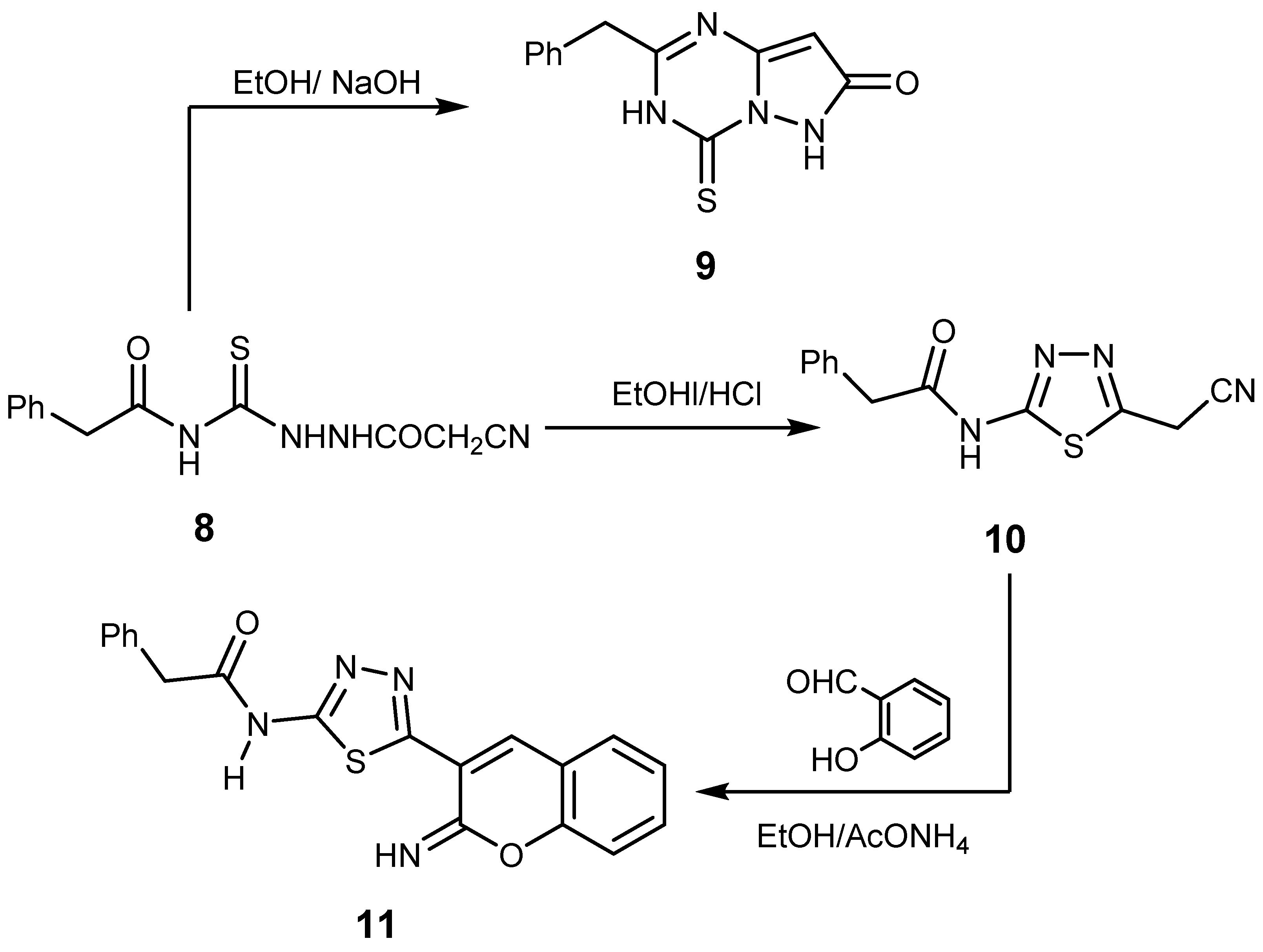
2.1. Chemistry
2.2. Biological Activity
2.2.1. Antimicrobial Sensitivity Assay
Structure–Activity Relationship (SAR)
2.2.2. Cytotoxic Activity Using MTT Assay
2.2.3. In Vitro Enzyme Assay
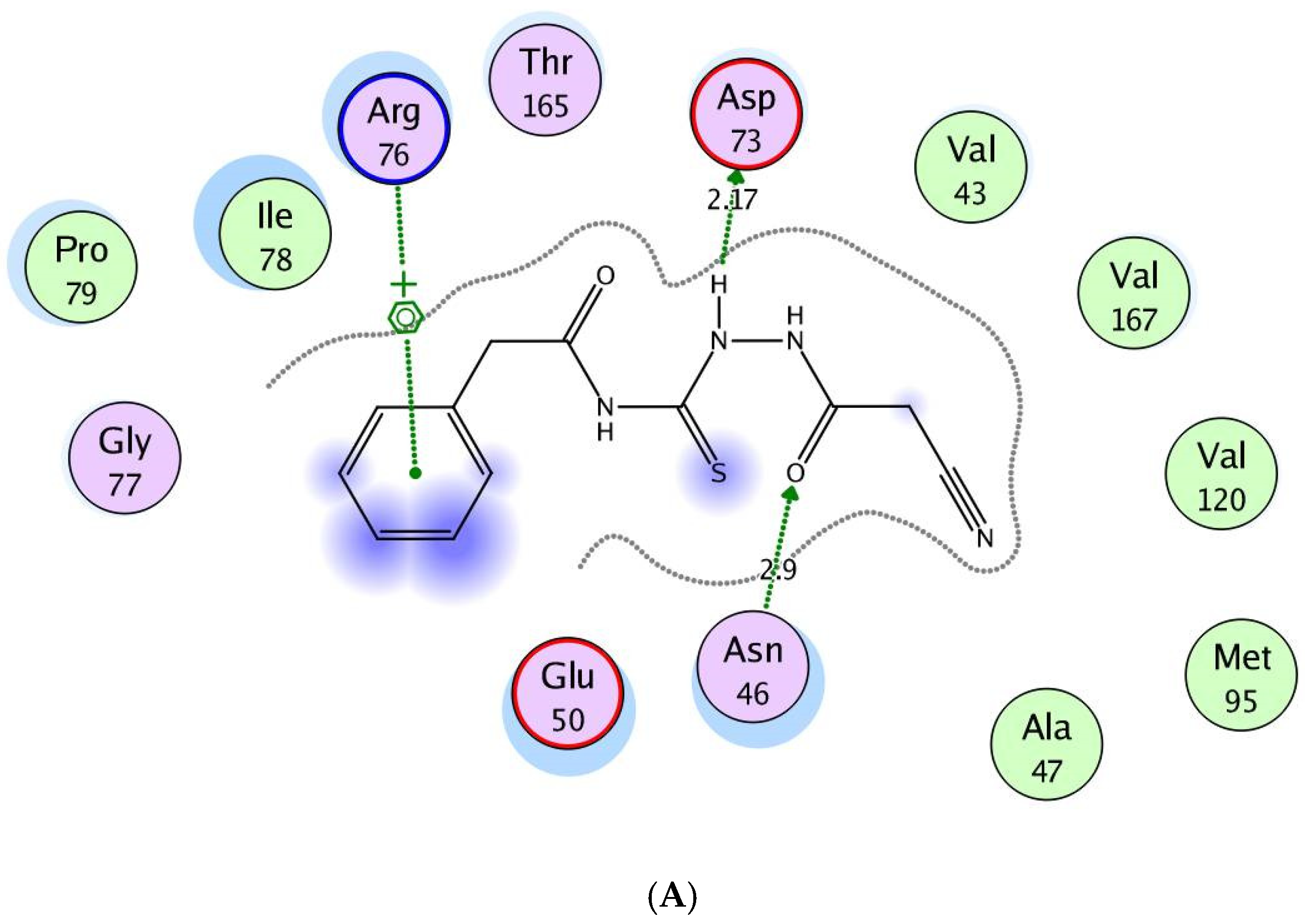
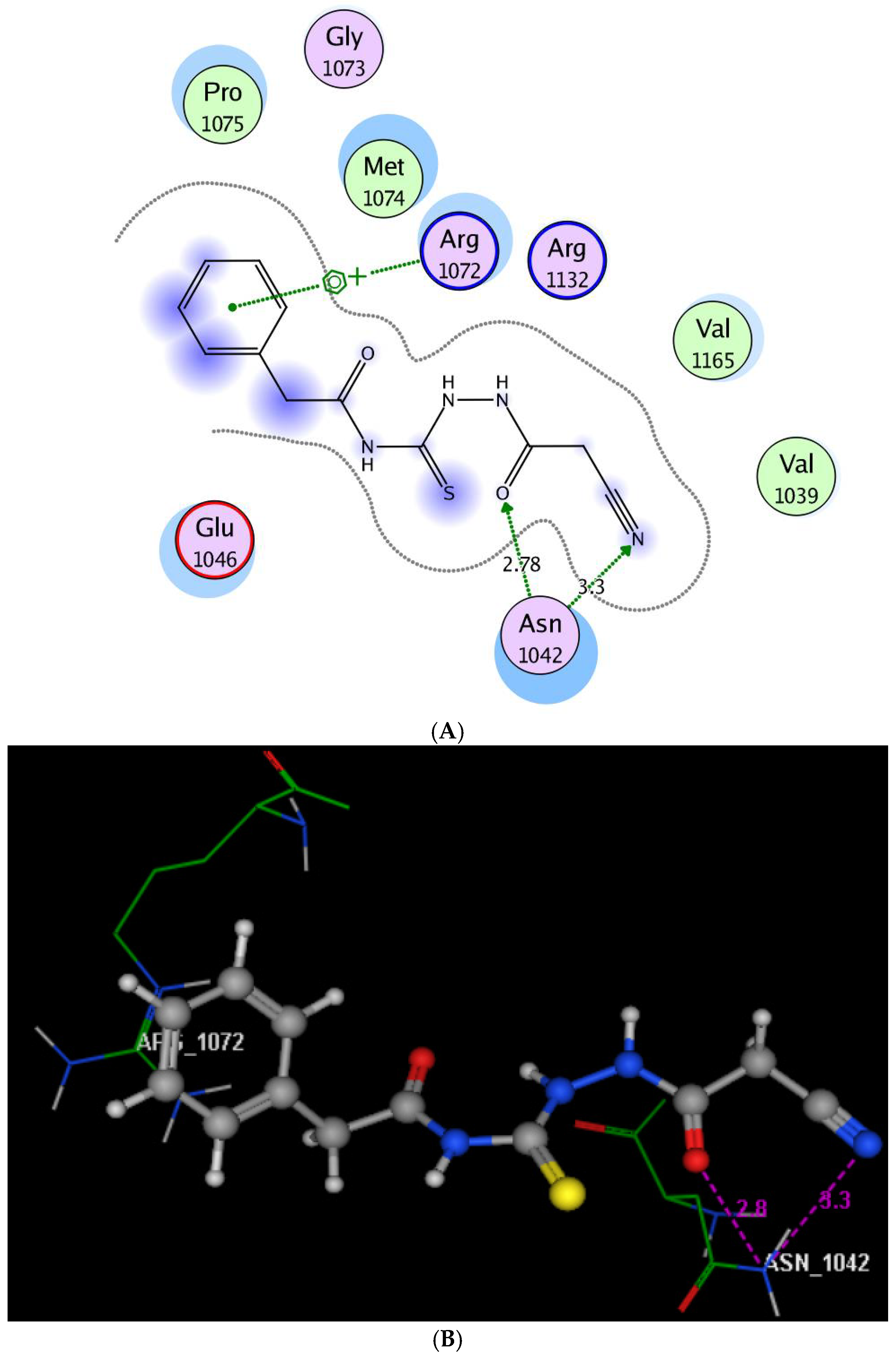
2.3. Molecular Modeling Study
3. Experimental
3.1. Chemistry
3.1.1. 3-(2-Phenylacetyl)-2-thioxoimidazolidin-4-one (3)
3.1.2. Ethyl ((2-phenylacetyl) carbono thionyl)glycinate (2)
3.1.3. Reactions of Isothiocyanate (1) with Different Amines. Synthesis of Compounds 6, 7 and 8
3.1.4. 1-(4-Benzylidene-4,5-dihydro-5-oxo-2-phenylimidazol-1-yl)-3-(2-phenylacetyl)thiourea (6)
3.1.5. 1-(2-Benzamido-3-phenylacryloyl)-4-phenylacetylthiosemicarbazide (7a)
3.1.6. 1-(2-Benzamido-3-(thiophen-2-yl)acryloyl)-4-phenylacetylthiosemicarbazide (7b)
3.1.7. 1-(2-Cyanoacetyl)-4-phenylacetylthiosemicarbazide (8)
3.1.8. Synthesis of Compounds 9 and 10
2-Benzyl-3,4-dihydro-4-thioxopyrazolo[1,5-a][1,3,5]triazin-7(6H)-one (9)
N-(5-(Cyanomethyl)-1,3,4-thiadiazol-2-yl)-2-phenylacetamide (10)
3.1.9. N-(5-(2-Imino-2H-chromen-3-yl)-1,3,4-thiadiazol-2-yl)-2-phenylacetamide (11)
3.1.10. 5-Benzylidene-5,6-dihydro-N-(1-hydroxy-2-phenylethylidene)-6-oxo-3-phenyl-1,2,4-triazine-2(1H)-carbothioamide (12)
3.1.11. N-(1-(5-(2-Phenylacetamido)-1,3,4-thiadiazol-2-yl)-2-(thiophen-2-yl)vinyl)benzamide (13)
3.2. Biological Activity
3.2.1. Antimicrobial Sensitivity Assay
3.2.2. MTT Assay for Cytotoxic Activity
3.2.3. Kinase Inhibition Assay
3.3. Molecular Modeling Study
4. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Fernandes, P.; Martens, E. Antibiotics in late clinical development. Biochem. Pharmacol. 2017, 133, 152–163. [Google Scholar] [CrossRef] [Green Version]
- Eakin, A.E.; Green, O.; Hales, N.; Walkup, G.K.; Bist, S.; Singh, A.; Mullen, G.; Bryant, J.; Embrey, K.; Gao, N.; et al. Pyrrolamide DNA gyrase inhibitors: Fragment-based nuclear magnetic resonance screening to identify antibacterial agents. Antimicrob. Agents Chemother. 2012, 56, 1240–1246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.; Wang, X.; Zhao, X.; Liang, Y.; He, H.; Fu, L. Synthesis and antitumor activity of novel quinazoline derivatives containing thiosemicarbazide moiety. Eur. J. Med. Chem. 2012, 54, 925–930. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Liu, H.; Fu, L.; Li, L.; Li, J.; Luo, X.; Mei, C. Antitumor activity of a new N-substituted thiourea derivative, an EGFR signaling-targeted inhibitor against a panel of human lung cancer cell lines. Chemotherapy 2008, 54, 463–474. [Google Scholar] [CrossRef]
- Pandeya, S.N.; Sriram, D.; Nath, G.; DeClercq, E. Synthesis, antibacterial, antifungal and anti-HIV activities of Schiff and Mannich bases derived from isatin derivatives and N-[4-(4′-chlorophenyl)thiazol-2-yl]thiosemicarbazide. Eur. J. Pharm. Sci. 1999, 9, 25–31. [Google Scholar] [CrossRef]
- Khan, S.A.; Singh, N.; Saleem, K. Synthesis, characterization and in vitro antibacterial activity of thiourea and urea derivatives of steroids. Eur. J. Med. Chem. 2008, 43, 2272–2277. [Google Scholar] [CrossRef] [PubMed]
- Bakherad, Z.; Mohammadi-Khanaposhtani, M.; Sadeghi-Aliabadi, H.; Rezaei, S.; Fassihi, A.; Bakherad, M.; Rastegar, H.; Biglar, M.; Saghaie, L.; Larijani, B.; et al. New thiosemicarbazide-1,2,3-triazole hybrids as potent a-glucosidase inhibitors: Design, synthesis, and biological evaluation. J. Mol. Struct. 2019, 1192, 192–200. [Google Scholar] [CrossRef]
- Ghorab, M.M.; El-Gaby, M.S.A.; Soliman, A.M.; Alsaid, M.S.; Abdel-Aziz, M.M.; Elaasser, M.M. Synthesis, docking study and biological evaluation of some new thiourea derivatives bearing benzenesulfonamide moiety. Chem. Cent. J. 2017, 11, 42. [Google Scholar] [CrossRef] [Green Version]
- Singhal, S.; Arora, S.; Agarwal, S.; Sharma, R.; Singhal, N. A review on potential biological activities of thiosemicarbazides. Wor. J. Pharm. Pharm. Sci. 2013, 2, 4661–4681. [Google Scholar]
- Galina, A.G.; Angelina, N.K. Thiosemicarbazides in the synthesis of five- and six membered heterocyclic compounds. Russ. Chem. Rev. 2012, 81, 494–523. [Google Scholar]
- Hassan, A.A.; Shawky, A.M. Thiosemicarbazides in heterocyclization. J. Hetero. Chem. 2011, 48, 495–516. [Google Scholar] [CrossRef]
- Ali, B.; Khan, K.M.; Salara, U.; Kanwal; Hussain, S.; Ashraf, M.; Riaz, M.; Wadood, A.; Taha, M.; Perveen, S. 1-[(4′-Chlorophenyl) carbonyl-4-(aryl) thiosemicarbazide derivatives as potent urease inhibitors: Synthesis, in vitro and in silico studies. Bioorg. Chem. 2018, 79, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Ukrainets, I.V.; Tkach, A.A.; Grinevich, L.A.; Turov, A.V.; Bevz, O.V. 4-Hydroxy-2-quinolones 165*. 1-R-4-hydroxy-2-oxo-1,2-dihydro-quinoline-3-carbaldehydes and their thiosemicarbazones. Synthesis, structure, and biological properties. Chem. Hetero. Comp. 2009, 45, 705–714. [Google Scholar] [CrossRef]
- Paneth, A.; Stączek, P.; Plech, T.; Strzelczyk, A.; Dzitko, K.; Wujec, M.; Kuśmierz, E.; Kosikowska, U.; Grzegorczyk, A.; Paneth, P. Biological evaluation and molecular modeling study of thiosemicarbazide derivatives as bacterial type IIA topoisomerases inhibitors. J. Enzym. Inhib. Med. Chem. 2016, 31, 14–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siwek, A.; Stączek, P.; Wujec, M.; Stefańska, J.; Kosikowska, U.; Malm, A.; Jankowski, S.; Paneth, P. Biological and docking studies of topoisomerase IV inhibition by thiosemicarbazides. J. Mol. Model. 2011, 17, 2297–2303. [Google Scholar] [CrossRef] [PubMed]
- Ghaly, M.A.; El-Bendary, E.R.; Shehata, I.A.; Bayomi, S.M.; Habib, E.E. Synthesis, antimicrobial activity, DNA-binding affinity and molecular docking of certain 1,2,4-triazolo[1,5-a]pyrimidines as nalidixic acid isosteres. J. Am. Sci. 2012, 8, 617–628. [Google Scholar]
- Bielenica, A.; Stefańska, J.; Stępień, K.; Napiórkowska, A.; Augustynowicz-Kopeć, E.; Sanna, G.; Madeddu, S.; Boi, S.; Giliberti, G.; Wrzosek, M.; et al. Synthesis, cytotoxicity and antimicrobial activity of thiourea derivatives incorporating 3-(trifluoromethyl)phenyl moiety. Eur. J. Med. Chem. 2015, 101, 111–125. [Google Scholar] [CrossRef]
- Hassan, G.S.; El-Messery, S.M.; Al-Omary, F.A.; Al-Rashood, S.T.; Shabayek, M.I.; Abulfadl, Y.S.; Habib, S.E.; El-Hallouty, S.M.; Fayad, W.; Mohamed, K.M.; et al. Nonclassical antifolates, part 4. 5-(2-aminothiazol-4-yl)-4-phenyl-4H-1,2,4-triazole-3-thiols as a new class of DHFR inhibitors: Synthesis, biological evaluation and molecular modeling study. Eur. J. Med. Chem. 2013, 66, 135–145. [Google Scholar] [CrossRef]
- Al-Omar, M.A.; Amr, A.E. Synthesis of some new pyridine-2,6-carboxamide-derived schiff bases as potential antimicrobial agents. Molecules 2010, 15, 4711–4721. [Google Scholar] [CrossRef]
- Khalifa, N.M.; Nossier, E.S.; Al-Omar, M.A.; Amr, A.E. Synthesis, reactions, and antimicrobial activity of some novel fused thiazolo[3,2-a]pyrimidine-5H-indeno[1,2-d]pyrimidine derivatives. Russ. J. Gen. Chem. 2016, 86, 1948–1953. [Google Scholar] [CrossRef]
- Khalifa, N.M.; El-Sayed, A.S.; Abd El-Karim, S.S.; Hassan, E.S.; Nossier, E.S.; Shalaby, A.G. 1,3,4-Triarylpyrazoles containing 2-thioxoimidazolidinones and different fused systems: Synthesis and antimicrobial activity. Russ. J. Gen. Chem. 2018, 88, 2646–2652. [Google Scholar] [CrossRef]
- El-Naggar, M.; Sallam, H.A.; Shaban, S.S.; Abdel-Wahab, S.S.; Amr, A.E.; Azab, M.E.; Nossier, E.S.; Al-Omar, M.A. Design, synthesis, and molecular docking study of novel heterocycles incorporating 1,3,4-thiadiazole moiety as potential antimicrobial and anticancer agents. Molecules 2019, 24, 1066. [Google Scholar] [CrossRef] [Green Version]
- El-Serwy, W.S.; Mohamed, H.S.; Mohamed, N.A.; Kassem, E.M.M.; Nossier, E.S.; Shalaby, A.G. Molecular docking study of newly synthesized thiopyrimidines as antimicrobial agents targeting DNA gyrase enzyme. J. Hetercycl. Chem. 2019, 56, 2027–2035. [Google Scholar] [CrossRef]
- Othman, I.M.M.; Gad-Elkareem, M.A.M.; El-Naggar, M.; Nossier, E.S.; Amr, A.E. Novel phthalimide based analogues: Design, synthesis, biological evaluation, and molecular docking studies. J. Enzym. Inhib. Med. Chem. 2019, 34, 1259–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassan, A.S.; Askar, A.A.; Nossier, E.S.; Naglah, A.M.; Moustafa, G.O.; Al-Omar, M.A. Antibacterial evaluation, in silico characters and molecular docking of Schiff Bases derived from 5-aminopyrazoles. Molecules 2019, 24, 3130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohi El-Deen, E.M.; Abd El-Meguid, E.A.; Hasabelnaby, S.; Karam, E.A.; Nossier, E.S. Synthesis, docking studies, and in vitro evaluation of some novel thienopyridines and fused thienopyridine-quinolines as antibacterial agents and DNA gyrase inhibitors. Molecules 2019, 24, 3650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lissi, E.; Modak, B.; Torres, R.; Escobar, J.; Urzúa, A. Total antioxidant potential of resinous exudates from Heliotropium species, and a comparison of the ABTS and DPPH methods. Free Radic. Res. 1999, 30, 471–477. [Google Scholar] [CrossRef]
- Deshmukh, M.B.; Chavan, P.B. Selective aziridination of olefins. Indian J. Chem. 1996, 35B, 1337–1339. [Google Scholar]
- Alt, S.; Mitchenall, L.A.; Maxwell, A.; Heide, L. Inhibition of DNA gyrase and DNA topoisomerase IV of Staphylococcus aureus and Escherichia coli by aminocoumarin antibiotics. J. Antimicrob. Chemother. 2011, 66, 2061–2069. [Google Scholar] [CrossRef] [Green Version]
- Elzahabi, H.S.A.; Nossier, E.S.; Khalifa, N.M.; Alasfoury, R.A.; El-Manawat, M.A. Anticancer evaluation and molecular modeling of multi-targeted kinase inhibitors based pyrido[2,3-d]pyrimidine scaffold. J. Enzym. Inhib. Med. Chem. 2018, 33, 546–557. [Google Scholar] [CrossRef] [Green Version]
- Bellon, S.; Parsons, J.D.; Wei, Y.; Hayakawa, K.; Swenson, L.L.; Charifson, P.S.; Lippke, J.A.; Aldape, R.; Gross, C.H. Crystal structures of Escherichia coli topoisomerase IV ParE subunit (24 and 43 Kilodaltons): A single residue dictates differences in novobiocin potency against topoisomerase IV and DNA gyrase. Antimicrob. Agents Chemother. 2004, 48, 1856–1864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Sample Availability: Samples of the compounds are not available from the authors. |







| Compd. | Mean Diameter of Inhibition Zone (Mean ± SEM) (mm) | |||||
|---|---|---|---|---|---|---|
| Gram Positive Bacteria | Gram Negative Bacteria | Fungi | ||||
| S. aureus ATCC 29213 | B. subtilis ATCC 6633 | P. aeruginosa ATCC 27853 | E. coli ATCC 25922 | A. flavus ATCC 46283 | C. albicans ATCC 10231 | |
| 2 | 9 ± 0.61 | NA | NA | NA | NA | NA |
| 3 | NA | NA | NA | NA | NA | NA |
| 6 | 27 ± 0.20 | 30 ± 0.26 | 21 ± 0.26 | 12 ± 0.10 | 9 ± 0.03 | 6 ± 0.22 |
| 7a | 30 ± 0.22 | 33 ± 0.50 | 27 ± 0.03 | 18 ± 0.22 | 24 ± 0.02 | 15 ± 0.30 |
| 7b | 33 ± 0.01 | 35 ± 0.26 | 26 ± 0.21 | 18 ± 0.41 | 25 ± 0.15 | 16 ± 0.12 |
| 8 | 39 ± 1.21 | 42 ± 0.30 | 36 ± 0.12 | 27 ± 0.62 | 27 ± 0.21 | 18 ± 0.05 |
| 9 | 21 ± 0.30 | 18 ± 0.25 | 9 ± 0.25 | NA | 15 ± 0.33 | 9 ± 0.20 |
| 10 | 15 ± 0.42 | 12 ± 0.21 | 6 ± 0.02 | NA | 6 ± 0.41 | NA |
| 11 | 12 ± 0.30 | 6 ± 0.53 | NA | NA | NA | NA |
| 12 | 22 ± 0.05 | 23 ± 0.01 | 17 ± 0.20 | 5 ± 0.58 | 18 ± 0.03 | 14 ± 0.21 |
| 13 | 21 ± 0.10 | 24 ± 1.11 | 15 ± 0.11 | 6 ± 0.15 | 18 ± 0.22 | 12 ± 0.46 |
| Ciprofloxacin | 24 ± 0.60 | 23 ± 0.20 | 23 ± 0.90 | 26 ± 0.25 | NA | NA |
| Clotrimazole | NA | NA | NA | NA | 25 ± 0.40 | 27 ± 0.21 |
| Compd. | MIC (Mean ± SEM) (μg/mL) | |||||
|---|---|---|---|---|---|---|
| Gram Positive Bacteria | Gram Negative Bacteria | Fungi | ||||
| S. aureus ATCC 29213 | B. subtilis ATCC 6633 | P. aeruginosa ATCC 27853 | E. coli ATCC 25922 | A. flavus ATCC 46283 | C. albicans ATCC 10231 | |
| 6 | 5.12 ± 0.05 | 2.29 ± 0.05 | 2.48 ± 0.11 | 7.80 ± 0.10 | 26.20 ± 0.03 | 91 ± 0.02 |
| 7a | 4.15 ± 0.22 | 1.39 ± 0.50 | 2.81 ± 0.01 | 4.22 ± 0.12 | 5.21 ± 0.11 | 64 ± 0.41 |
| 7b | 3.90 ± 0.26 | 1.46 ± 0.10 | 2.72 ± 0.15 | 4.68 ± 0.13 | 5.70 ± 0.01 | 76 ± 0.31 |
| 8 | 3.25 ± 1.00 | 1.38 ± 0.25 | 1.25 ± 0.50 | 0.95 ± 0.22 | 2.11 ± 0.51 | 39.8 ± 0.20 |
| 9 | 9.92 ± 0.30 | 8.60 ± 0.25 | 17.62 ± 0.20 | - | 20.45 ± 0.30 | 94 ± 0.10 |
| 12 | 5.22 ± 0.03 | 2.30 ± 0.05 | 12.50 ± 0.20 | 27 ± 0.02 | 15.60 ± 0.01 | 85 ± 0.15 |
| 13 | 5.60 ± 0.15 | 2.72 ± 1.11 | 13.41 ± 0.10 | 33. ± 0.10 | 14.55 ± 0.20 | 83 ± 0.22 |
| Ciprofloxacin | 5.85 ± 0.13 | 2.90 ± 0.02 | 2.90 ± 0.04 | 2.90 ± 0.25 | - | - |
| Clotrimazole | - | - | - | - | 4.25 ± 0.05 | 12.5 ± 0.15 |
| Compd. No. | IC50 (Mean ± SEM) (µM) a | IC50 (Mean ± SEM) (µM) a |
|---|---|---|
| MCF-7 Cells | Vero Cells | |
| Cisplatin | 8.897 ± 0.37 | 92.16 ± 0.07 |
| 6 | 23.69 ± 0.96 | 122.81 ± 0.40 |
| 7a | 10.17 ± 0.65 | 149.10 ± 0.21 |
| 7b | 11.589 ± 0.59 | 133.26 ± 0.40 |
| 8 | 22.35 ± 0.36 | 144.72 ± 0.36 |
| 9 | 26.45 ± 0.46 | 62.45 ± 0.20 |
| 12 | 35.92 ± 0.85 | 79.19 ± 0.28 |
| 13 | 37.68 ± 0.93 | 84.11 ± 0.32 |
| Kinase | IC50 (mean ± SEM) (µM) | ||
|---|---|---|---|
| 8 | Novobiocin | Methotrexate | |
| DNA gyrase B | 0.33 ± 1.25 | 0.28 ± 1.45 | - |
| DNA topoisomerase IV | 19.72 ± 1.00 | 10.65 ± 1.02 | - |
| DHFR | 189.47 ± 1.06 | - | 0.14 ± 1.62 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hashem, H.E.; Amr, A.E.-G.E.; Nossier, E.S.; Elsayed, E.A.; Azmy, E.M. Synthesis, Antimicrobial Activity and Molecular Docking of Novel Thiourea Derivatives Tagged with Thiadiazole, Imidazole and Triazine Moieties as Potential DNA Gyrase and Topoisomerase IV Inhibitors. Molecules 2020, 25, 2766. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules25122766
Hashem HE, Amr AE-GE, Nossier ES, Elsayed EA, Azmy EM. Synthesis, Antimicrobial Activity and Molecular Docking of Novel Thiourea Derivatives Tagged with Thiadiazole, Imidazole and Triazine Moieties as Potential DNA Gyrase and Topoisomerase IV Inhibitors. Molecules. 2020; 25(12):2766. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules25122766
Chicago/Turabian StyleHashem, Heba E., Abd El-Galil E. Amr, Eman S. Nossier, Elsayed A. Elsayed, and Eman M. Azmy. 2020. "Synthesis, Antimicrobial Activity and Molecular Docking of Novel Thiourea Derivatives Tagged with Thiadiazole, Imidazole and Triazine Moieties as Potential DNA Gyrase and Topoisomerase IV Inhibitors" Molecules 25, no. 12: 2766. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules25122766








