Current Advances in the Bacterial Toolbox for the Biotechnological Production of Monoterpene-Based Aroma Compounds
Abstract
:1. The Importance of Aroma Compounds in Industry
1.1. Scope of the Review
1.2. Aroma Compounds in Nature: Monoterpenes and Monoterpenoids
2. Catalysis Mediated by Biological Systems
2.1. Bacterial Adaptation to the Hydrophobicity of Monoterpene Substrates
- I.
- II.
- III.
- They may alter the profile of phospholipid head groups, which is predicted to influence the physical and chemical properties of the membrane (e.g., charge and melting point) [31].
- IV.
- They may swiftly increase cell surface hydrophobicity by altering the composition of the lipopolysaccharide layer (e.g., complete loss of B band lipopolysaccharide), and generate outer membrane vesicles (OMVs). Although this ubiquitous mechanism has not been extensively reported as a response to hydrophobic stress, several studies with solvent-tolerant P. putida strains have shown the induction of vesiculation mediated by alkanes and alkanols ([36] and references therein). This strategy may provide an enhanced ability for protective cell attachment, aggregation and biofilm formation, as well as for partitioning the hydrocarbon stressor in vesicles ([37] and references therein).
- V.
- Several bacterial strains (e.g., Pseudomonas spp., Vibrio spp., strains of Methylococcus capsulatus, Alcanivorax borkumensis and Colwellia psychrerythraea) employ a fifth adaptive mechanism by isomerizing cis-unsaturated fatty acids to trans-unsaturated acyl chains ([37] and references therein). Cis-unsaturated acyl chains comprise a bend of 30°, which disturbs the ordered fatty acid packing, increasing fluidity, allowing denser packing and promoting an increase in membrane stiffness to counteract excessive fluidity ([37] and references therein). This membrane cis-to-trans isomerization is performed by cis/trans isomerases, and since it is dependent on neither energy nor on the de novo synthesis of fatty acid molecules, this mechanism is considered a rapid short-term response to chemical stress.
2.2. Mechanisms for the Bacterial Transformation of the Hydrocarbon Backbone
2.2.1. Molecular Mechanism for the Catabolism of the Unsaturated Hydrocarbon Backbone
2.2.2. Molecular Mechanism for the Catabolism of the Branched Hydrocarbon Backbone
2.3. Nature’s Reservoir of Bacterial Biocatalysts for Industrially Relevant Monoterpenes
2.3.1. Pinene Isomers: A Bicyclic Precursor
2.3.2. Limonene: The Monocyclic Precursor
2.3.3. β-Myrcene: The Versatile Acyclic Precursor
3. Exploiting the Biotechnological Potential of Monoterpene-Catabolizing Enzymes
3.1. Metagenomics Approaches May Expand the Bacterial Toolbox for the Production of Monoterpene-Based Aroma Compounds
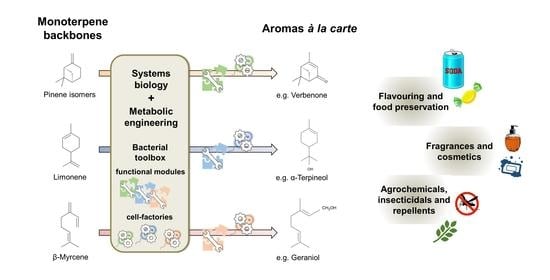
3.2. Holistic Approaches Are the Framework for Monoterpene Biocatalysis À La Carte
3.3. Coupling the Bacterial Synthesis of Monoterpene Precursors with the Oxidative Biocatalysis into Aroma Compounds
4. Outlook and Final Considerations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dehsheikh, A.B.; Sourestani, M.M.; Dehsheikh, P.B.; Mottaghipisheh, J.; Vitalini, S.; Iriti, M. Monoterpenes: Essential oil components with valuable features. Mini Rev. Med. Chem. 2020, 20, 958–974. [Google Scholar] [CrossRef] [PubMed]
- Tsolakis, N.; Bam, W.; Srai, J.S.; Kumar, M. Renewable chemical feedstock supply network design: The case of terpenes. J. Clean. Prod. 2019, 222, 802–822. [Google Scholar] [CrossRef]
- Jiang, Z.; Kempinski, C.; Chappell, J. Extraction and analysis of terpenes/terpenoids. Curr. Protoc. Plant Biol. 2016, 1, 345–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Oliveira Felipe, L.; de Oliveira, A.M.; Bicas, J.L. Bioaromas—Perspectives for sustainable development. Trends Food Sci. Technol. 2017, 62, 141–153. [Google Scholar] [CrossRef]
- Longo, M.A.; Sanromán, M.A. Production of food aroma compounds: Microbial and enzymatic methodologies. Food Technol. Biotechnol. 2006, 44, 335–353. [Google Scholar]
- Sheldon, R.A.; Woodley, J.M. Role of biocatalysis in sustainable chemistry. Chem. Rev. 2018, 118, 801–838. [Google Scholar] [CrossRef]
- Krings, U.; Berger, G.R. Biotechnological production of flavours and fragrances. Appl. Microbiol. Biotechnol. 1998, 49, 1–8. [Google Scholar] [CrossRef]
- Hatti-Kaul, R.; Törnvall, U.; Gustafsson, L.; Börjesson, P. Industrial biotechnology for the production of bio-based chemicals—A cradle-to-grave perspective. Trends Biotechnol. 2007, 25, 119–124. [Google Scholar] [CrossRef]
- Li, C.-J.; Trost, B.M. Green chemistry for chemical synthesis. Proc. Natl. Acad. Sci. USA 2008, 105, 13197–13202. [Google Scholar] [CrossRef] [Green Version]
- European Commission. Regulation (EC) No 1334/2008. Off. J. Eur. Union L 2008, 354, 34–50. [Google Scholar]
- Food Drug Administration. A Food Labeling Guide; USA Food Drug Administration: Silver Spring, MD, USA, 2013; Volume 132.
- Dionísio, A.P.; Molina, G.; de Carvalho, D.S.; Dos Santos, R.; Bicas, J.L.; Pastore, G.M. Natural flavourings from biotechnology for foods and beverages. In Natural Food Additives, Ingredients and Flavourings; Woodhead Publishing: Cambridge, UK, 2012; pp. 231–259. [Google Scholar] [CrossRef]
- Burdock, G.A. Fenaroli’s Handbook of Flavor Ingredients, 6th ed.; CRC Press: Boca Raton, FL, USA, 2019; Volume 2, pp. 1–2035. ISBN 1000694666. [Google Scholar]
- Schempp, F.M.; Drummond, L.; Buchhaupt, M.; Schrader, J. Microbial cell factories for the production of terpenoid flavor and fragrance compounds. J. Agric. Food Chem. 2017, 66, 2247–2258. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, G.; Chattopadhyay, P. Vanillin biotechnology: The perspectives and future. J. Sci. Food Agric. 2019, 99, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Caputi, L.; Aprea, E. Use of terpenoids as natural flavouring compounds in food industry. Recent Pat. Food Nutr. Agric. 2011, 3, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Paulino, B.N.; Sales, A.; de Oliveira Felipe, L.; Pastore, G.M.; Molina, G.; Bicas, J.L. Recent advances in the microbial and enzymatic production of aroma compounds. Curr. Opin. Food Sci. 2021, 37, 98–106. [Google Scholar] [CrossRef]
- Sales, A.; Paulino, B.N.; Pastore, G.M.; Bicas, J.L. Biogeneration of aroma compounds. Curr. Opin. Food Sci. 2018, 19, 77–84. [Google Scholar] [CrossRef]
- Braga, A.; Guerreiro, C.; Belo, I. Generation of flavors and fragrances through biotransformation and de novo synthesis. Food Bioprocess. Technol. 2018, 11, 2217–2228. [Google Scholar] [CrossRef] [Green Version]
- Soares, G.P.A.; Souza, K.S.T.; Vilela, L.F.; Schwan, R.F.; Dias, D.R. γ-Decalactone production by Yarrowia lipolytica and Lindnera saturnus in crude glycerol. Prep. Biochem. Biotechnol. 2017, 47, 633–637. [Google Scholar] [CrossRef]
- Hansen, E.H.; Møller, B.L.; Kock, G.R.; Bünner, C.M.; Kristensen, C.; Jensen, O.R.; Okkels, F.T.; Olsen, C.E.; Motawia, M.S.; Hansen, J. De novo biosynthesis of vanillin in fission yeast (Schizosaccharomyces pombe) and baker’s yeast (Saccharomyces cerevisiae). Appl. Environ. Microbiol. 2009, 75, 2765–2774. [Google Scholar] [CrossRef] [Green Version]
- Tholl, D. Biosynthesis and biological functions of terpenoids in plants. In Biotechnology of Isoprenoids; Springer: Cham, Switzerland, 2015; pp. 63–106. [Google Scholar] [CrossRef]
- Langenheim, J.H. Higher plant terpenoids: A phytocentric overview of their ecological roles. J. Chem. Ecol. 1994, 20, 1223–1280. [Google Scholar] [CrossRef]
- Schwab, W.; Fuchs, C.; Huang, F.-C. Transformation of terpenes into fine chemicals. Eur. J. Lipid Sci. Technol. 2013, 115, 3–8. [Google Scholar] [CrossRef]
- Adams, T.B.; Gavin, C.L.; McGowen, M.M.; Waddell, W.J.; Cohen, S.M.; Feron, V.J.; Marnett, L.J.; Munro, I.C.; Portoghese, P.S.; Rietjens, I. The FEMA GRAS assessment of aliphatic and aromatic terpene hydrocarbons used as flavor ingredients. Food Chem. Toxicol. 2011, 49, 2471–2494. [Google Scholar] [CrossRef] [PubMed]
- Brenner, K.; You, L.; Arnold, F.H. Engineering microbial consortia: A new frontier in synthetic biology. Trends Biotechnol. 2008, 26, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Gavrilescu, M.; Chisti, Y. Biotechnology—a sustainable alternative for chemical industry. Biotechnol. Adv. 2005, 23, 471–499. [Google Scholar] [CrossRef] [PubMed]
- Sikkema, J.; de Bont, J.A.; Poolman, B. Mechanisms of membrane toxicity of hydrocarbons. Microbiol. Rev. 1995, 59, 201–222. [Google Scholar] [CrossRef] [PubMed]
- Del Turina, A.V.; Nolan, M.V.; Zygadlo, J.A.; Perillo, M.A. Natural terpenes: Self-assembly and membrane partitioning. Biophys. Chem. 2006, 122, 101–113. [Google Scholar] [CrossRef]
- Soares-Castro, P.; Montenegro-Silva, P.; Heipieper, H.J.; Santos, P.M. Functional characterization of a 28-kilobase catabolic island from Pseudomonas sp. strain M1 involved in biotransformation of β-myrcene and related plant-derived volatiles. Appl. Environ. Microbiol. 2017, 83, e03112–e03116. [Google Scholar] [CrossRef] [Green Version]
- Ramos, J.L.; Duque, E.; Gallegos, M.-T.; Godoy, P.; Ramos-González, M.I.; Rojas, A.; Terán, W.; Segura, A. Mechanisms of solvent tolerance in Gram-negative bacteria. Annu. Rev. Microbiol. 2002, 56, 743–768. [Google Scholar] [CrossRef]
- Murínová, S.; Dercová, K. Response mechanisms of bacterial degraders to environmental contaminants on the level of cell walls and cytoplasmic membrane. Int. J. Microbiol. 2014, 873081, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Di Pasqua, R.; Hoskins, N.; Betts, G.; Mauriello, G. Changes in membrane fatty acids composition of microbial cells induced by addiction of thymol, carvacrol, limonene, cinnamaldehyde, and eugenol in the growing media. J. Agric. Food Chem. 2006, 54, 2745–2749. [Google Scholar] [CrossRef]
- De Carvalho, C.C.C.R.; Parreño-Marchante, B.; Neumann, G.; da Fonseca, M.M.R.; Heipieper, H.J. Adaptation of Rhodococcus erythropolis DCL14 to growth on n-alkanes, alcohols and terpenes. Appl. Microbiol. Biotechnol. 2005, 67, 383–388. [Google Scholar] [CrossRef]
- Kabelitz, N.; Santos, P.M.; Heipieper, H.J. Effect of aliphatic alcohols on growth and degree of saturation of membrane lipids in Acinetobacter calcoaceticus. FEMS Microbiol. Lett. 2003, 220, 223–227. [Google Scholar] [CrossRef] [Green Version]
- Eberlein, C.; Starke, S.; Doncel, Á.E.; Scarabotti, F.; Heipieper, H.J. Quantification of outer membrane vesicles: A potential tool to compare response in Pseudomonas putida KT2440 to stress caused by alkanols. Appl. Microbiol. Biotechnol. 2019, 103, 4193–4201. [Google Scholar] [CrossRef] [PubMed]
- Eberlein, C.; Baumgarten, T.; Starke, S.; Heipieper, H.J. Immediate response mechanisms of Gram-negative solvent-tolerant bacteria to cope with environmental stress: Cis-trans isomerization of unsaturated fatty acids and outer membrane vesicle secretion. Appl. Microbiol. Biotechnol. 2018, 102, 2583–2593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Beilen, J.B.; Funhoff, E.G. Alkane hydroxylases involved in microbial alkane degradation. Appl. Microbiol. Biotechnol. 2007, 74, 13–21. [Google Scholar] [CrossRef] [Green Version]
- Fujita, Y.; Matsuoka, H.; Hirooka, K. Regulation of fatty acid metabolism in bacteria. Mol. Microbiol. 2007, 66, 829–839. [Google Scholar] [CrossRef]
- Ji, Y.; Mao, G.; Wang, Y.; Bartlam, M. Structural insights into diversity and n-alkane biodegradation mechanisms of alkane hydroxylases. Front. Microbiol. 2013, 4, 58. [Google Scholar] [CrossRef] [Green Version]
- Abbasian, F.; Lockington, R.; Mallavarapu, M.; Naidu, R. A comprehensive review of aliphatic hydrocarbon biodegradation by bacteria. Appl. Biochem. Biotechnol. 2015, 176, 670–699. [Google Scholar] [CrossRef]
- Washisu, S.; Aida, T.; Hashimoto, N.; Kanisawa, T. Production of Vanillin and Its Related Compound by Fermentation. Patent Application JP5227980, 7 September 1993. [Google Scholar]
- Labuda, I.M.; Goers, S.K.; Keon, K.A. Bioconversion Process for the Production of Vanillin. U.S. Patent No. 5128253, 7 July 1992. [Google Scholar]
- Rhodes, P.M.; Winskill, N. Microbiological Process for the Preparation of 1-Carvone. U.S. Patent No. 4495284, 22 January 1985. [Google Scholar]
- Brady, D.; Reddy, S.; Mboniswa, B.; Steenkamp, L.H.; Rousseau, A.L.; Parkinson, C.J.; Chaplin, J.; Mitra, R.K.; Moutlana, T.; Marais, S.F.; et al. Biocatalytic enantiomeric resolution of l-menthol from an eight isomeric menthol mixture through transesterification. J. Mol. Catal. B Enzym. 2012, 75, 1–10. [Google Scholar] [CrossRef]
- Serra, S.; Brenna, E.; Fuganti, C.; Maggioni, F. Lipase-catalyzed resolution of p-menthan-3-ols monoterpenes: Preparation of the enantiomer-enriched forms of menthol, isopulegol, trans- and cis-piperitol, and cis-isopiperitenol. Tetrahedron Asymmetry 2003, 14, 3313–3319. [Google Scholar] [CrossRef]
- Zheng, G.; Yu, H.; Zhang, J.; Xu, J. Enzymatic production of L-menthol by a high substrate concentration tolerable esterase from newly isolated Bacillus subtilis ECU0554. Adv. Synth. Catal. 2009, 351, 405–414. [Google Scholar] [CrossRef]
- Van Beilen, J.B.; Wubbolts, M.G.; Witholt, B. Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation 1994, 5, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Van Beilen, J.B.; Panke, S.; Lucchini, S.; Franchini, A.G.; Röthlisberger, M.; Witholt, B. Analysis of Pseudomonas putida alkane-degradation gene clusters and flanking insertion sequences: Evolution and regulation of the alk genes. Microbiology 2001, 147, 1621–1630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheps, D.; Malca, S.H.; Hoffmann, H.; Nestl, B.M.; Hauer, B. Regioselective ω-hydroxylation of medium-chain n-alkanes and primary alcohols by CYP153 enzymes from Mycobacterium marinum and Polaromonas sp. strain JS666. Org. Biomol. Chem. 2011, 9, 6727–6733. [Google Scholar] [CrossRef] [PubMed]
- Maeng, J.H.; Sakai, Y.; Tani, Y.; Kato, N. Isolation and characterization of a novel oxygenase that catalyzes the first step of n-alkane oxidation in Acinetobacter sp. strain M-1. J. Bacteriol. 1996, 178, 3695–3700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, W.J.; Puah, S.M.; Tan, L.L.; Ng, S.S. Production of (R)-ethyl-3,4-epoxybutyrate by newly isolated Acinetobacter baumannii containing epoxide hydrolase. Appl. Microbiol. Biotechnol. 2008, 79, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Van der Werf, M.J.; Overkamp, K.M.; de Bont, J.A.M. Limonene-1, 2-epoxide hydrolase from Rhodococcus erythropolis DCL14 belongs to a novel class of epoxide hydrolases. J. Bacteriol. 1998, 180, 5052–5057. [Google Scholar] [CrossRef] [Green Version]
- Van Hylckama Vlieg, J.E.T.; Kingma, J.; Kruizinga, W.; Janssen, D.B. Purification of a glutathione s-transferase and a glutathione conjugate-specific dehydrogenase involved in isoprene metabolism in Rhodococcus sp. strain AD45. J. Bacteriol. 1999, 181, 2094–2101. [Google Scholar] [CrossRef] [Green Version]
- Weijers, C.A.G.M.; de Haan, A.; de Bont, J.A.M. Chiral resolution of 2,3-epoxyalkanes by Xanthobacter Py2. Appl. Microbiol. Biotechnol. 1988, 27, 337–340. [Google Scholar] [CrossRef]
- Leisch, H.; Morley, K.; Lau, P.C.K. Baeyer-Villiger monooxygenases: More than just green chemistry. Chem. Rev. 2011, 111, 4165–4222. [Google Scholar] [CrossRef] [Green Version]
- Grossi, V.; Cravo-Laureau, C.; Guyoneaud, R.; Ranchou-Peyruse, A.; Hirschler-Réa, A. Metabolism of n-alkanes and n-alkenes by anaerobic bacteria: A summary. Org. Geochem. 2008, 39, 1197–1203. [Google Scholar] [CrossRef]
- Brodkorb, D.; Gottschall, M.; Marmulla, R.; Lüddeke, F.; Harder, J. Linalool dehydratase-isomerase, a bifunctional enzyme in the anaerobic degradation of monoterpenes. J. Biol. Chem. 2010, 285, 30436–30442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkes, H.; Buckel, W.; Golding, B.T.; Rabus, R. Metabolism of hydrocarbons in n-alkane-utilizing anaerobic bacteria. J. Mol. Microbiol. Biotechnol. 2016, 26, 138–151. [Google Scholar] [CrossRef] [PubMed]
- Kung, J.W.; Meier, A.-K.; Mergelsberg, M.; Boll, M. Enzymes involved in a novel anaerobic cyclohexane carboxylic acid degradation pathway. J. Bacteriol. 2014, 196, 3667–3674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaeffer, T.L.; Cantwell, S.G.; Brown, J.L.; Watt, D.S.; Fall, R.R. Microbial growth on hydrocarbons: Terminal branching inhibits biodegradation. Appl. Environ. Microbiol. 1979, 38, 742–746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pirnik, M.P.; McKenna, E.J. Microbial oxidation of methyl branched alkanes. CRC Crit. Rev. Microbiol. 1977, 5, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Cantwell, S.G.; Lau, E.P.; Watt, D.S.; Fall, R.R. Biodegradation of acyclic isoprenoids by Pseudomonas species. J. Bacteriol. 1978, 135, 324–333. [Google Scholar] [CrossRef] [Green Version]
- Marmulla, R.; Cala, E.P.; Markert, S.; Schweder, T.; Harder, J. The anaerobic linalool metabolism in Thauera linaloolentis 47 Lol. BMC Microbiol. 2016, 16, 76. [Google Scholar] [CrossRef] [Green Version]
- Förster-Fromme, K.; Jendrossek, D. Identification and characterization of the acyclic terpene utilization gene cluster of Pseudomonas citronellolis. FEMS Microbiol. Lett. 2006, 264, 220–225. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Liu, Y.-F.; Zhou, L.; Mbadinga, S.M.; Yang, T.; Zhou, J.; Liu, J.-F.; Yang, S.-Z.; Gu, J.-D.; Mu, B.-Z. Methanogenic degradation of branched alkanes in enrichment cultures of production water from a high-temperature petroleum reservoir. Appl. Microbiol. Biotechnol. 2019, 103, 2391–2401. [Google Scholar] [CrossRef]
- Vespermann, K.A.C.; Paulino, B.N.; Barcelos, M.C.S.; Pessôa, M.G.; Pastore, G.M.; Molina, G. Biotransformation of α-and β-pinene into flavor compounds. Appl. Microbiol. Biotechnol. 2017, 101, 1805–1817. [Google Scholar] [CrossRef]
- Behr, A.; Johnen, L. Myrcene as a natural base chemical in sustainable chemistry: A critical review. Chem. Sus. Chem. 2009, 2, 1072–1095. [Google Scholar] [CrossRef] [PubMed]
- Bicas, J.L.; Dionísio, A.P.; Pastore, G.M. Bio-oxidation of terpenes: An approach for the flavor industry. Chem. Rev. 2009, 109, 4518–4531. [Google Scholar] [CrossRef] [PubMed]
- Savithiry, N.; Gage, D.; Fu, W.; Oriel, P. Degradation of pinene by Bacillus pallidus BR425. Biodegradation 1998, 9, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Tudroszen, N.J.; Kelly, D.P.; Millis, N.F. α-Pinene metabolism by Pseudomonas putida. Biochem. J. 1977, 168, 315–318. [Google Scholar] [CrossRef]
- Fontanille, P.; Le Fleche, A.; Larroche, C. Pseudomonas rhodesiae PF1: A new and efficient biocatalyst for production of isonovalal from α-pinene oxide. Biocatal. Biotransform. 2002, 20, 413–421. [Google Scholar] [CrossRef]
- Gibbon, G.H.; Pirt, S.J. The degradation of α-pinene by Pseudomonas PX1. FEBS Lett. 1971, 18, 103–105. [Google Scholar] [CrossRef] [Green Version]
- Bicas, J.L.; Fontanille, P.; Pastore, G.M.; Larroche, C. Characterization of monoterpene biotransformation in two pseudomonads. J. Appl. Microbiol. 2008, 105, 1991–2001. [Google Scholar] [CrossRef]
- Griffiths, E.T.; Harries, P.C.; Jeffcoat, R.; Trudgill, P.W. Purification and properties of alpha-pinene oxide lyase from Nocardia sp. strain P18.3. J. Bacteriol. 1987, 169, 4980–4983. [Google Scholar] [CrossRef] [Green Version]
- Yoo, S.K.; Day, D.F. Bacterial metabolism of α- and β-pinene and related monoterpenes by Pseudomonas sp. strain PIN. Process. Biochem. 2002, 37, 739–745. [Google Scholar] [CrossRef]
- Cheng, Z.; Sun, P.; Jiang, Y.; Zhang, L.; Chen, J. Kinetic analysis and bacterium metabolization of α-pinene by a novel identified Pseudomonas sp. strain. J. Environ. Sci. 2012, 24, 1806–1815. [Google Scholar] [CrossRef]
- Colocousi, A.; Saqib, M.K.; Leak, J.D. Mutants of Pseudomonas fluorescens NCIMB 11671 defective in the catabolism of α-pinene. Appl. Microbiol. Biotechnol. 1996, 45, 822–830. [Google Scholar] [CrossRef]
- Wright, S.J.; Caunt, P.; Carter, D.; Baker, P.B. Microbial oxidation of alpha-pinene by Serratia marcescens. Appl. Microbiol. Biotechnol. 1986, 23, 224–227. [Google Scholar] [CrossRef]
- Best, D.J.; Floyd, N.C.; Magalhaes, A.; Burfield, A.; Rhodes, P.M. Initial enzymatic steps in the degradation of alpha-pinene by Pseudomonas fluorescens NCIMB 11671. Biocatalysis 1987, 1, 147–159. [Google Scholar] [CrossRef]
- Fontanille, P.; Larroche, C. Optimization of isonovalal production from α-pinene oxide using permeabilized cells of Pseudomonas rhodesiae CIP 107491. Appl. Microbiol. Biotechnol. 2003, 60, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Dubessay, P.; Larroche, C.; Fontanille, P. Cloning and Characterization of the gene encoding alpha-pinene oxide lyase enzyme (Prα-POL) from Pseudomonas rhodesiae CIP 107491 and production of the recombinant protein in Escherichia coli. Appl. Biochem. Biotechnol. 2018, 185, 676–690. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Liu, S.; Jin, G.; Yang, X.; Zhou, Y.J. Microbial production of limonene and its derivatives: Achievements and perspectives. Biotechnol. Adv. 2020, 44, 107628. [Google Scholar] [CrossRef]
- Ciriminna, R.; Lomeli-Rodriguez, M.; Demma Cara, P.; Lopez-Sanchez, J.A.; Pagliaro, M. Limonene: A versatile chemical of the bioeconomy. Chem. Commun. 2014, 50, 15288–15296. [Google Scholar] [CrossRef]
- Van der Werf, M.J.; Orru, R.V.A.; Overkamp, K.M.; Swarts, H.J.; Osprian, I.; Steinreiber, A.; de Bont, J.A.M.; Faber, K. Substrate specificity and stereospecificity of limonene-1,2-epoxide hydrolase from Rhodococcus erythropolis DCL14; an enzyme showing sequential and enantioconvergent substrate conversion. Appl. Microbiol. Biotechnol. 1999, 52, 380–385. [Google Scholar] [CrossRef]
- Van der Werf, M.J.; Swarts, H.J.; de Bont, J.A.M. Rhodococcus erythropolis DCL14 contains a novel degradation pathway for limonene. Appl. Environ. Microbiol. 1999, 65, 2092–2102. [Google Scholar] [CrossRef] [Green Version]
- Maga, J.A.; Katz, I. Lactones in foods. Crit. Rev. Food Sci. Nutr. 1976, 8, 1–56. [Google Scholar] [CrossRef]
- Duetz, W.A.; Fjällman, A.H.M.; Ren, S.; Jourdat, C.; Witholt, B. Biotransformation of D-limonene to (+)-trans-carveol by toluene-grown Rhodococcus opacus PWD4 cells. Appl. Environ. Microbiol. 2001, 67, 2829–2832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petasch, J.; Disch, E.-M.; Markert, S.; Becher, D.; Schweder, T.; Hüttel, B.; Reinhardt, R.; Harder, J. The oxygen-independent metabolism of cyclic monoterpenes in Castellaniella defragrans 65Phen. BMC Microbiol. 2014, 14, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, H.C.; Oriel, P. Bioproduction of perillyl alcohol and related monoterpenes by isolates of Bacillus stearothermophilus. J. Food Sci. 1994, 59, 660–662. [Google Scholar] [CrossRef]
- Park, Y.J.; Kim, I.C.; Chang, H.C. Microbial conversion of (+)-limonene by an Enterobacter agglomerans isolate. J. Microbiol. Biotechnol. 2003, 13, 636–639. [Google Scholar]
- Mirata, M.A.; Heerd, D.; Schrader, J. Integrated bioprocess for the oxidation of limonene to perillic acid with Pseudomonas putida DSM 12264. Process. Biochem. 2009, 44, 764–771. [Google Scholar] [CrossRef]
- Chatterjee, T.; Bhattacharyya, D. Biotransformation of limonene by Pseudomonas putida. Appl. Microbiol. Biotechnol. 2001, 55, 541–546. [Google Scholar] [CrossRef]
- Van der Werf, M.J.; Keijzer, P.M.; van der Schaft, P.H. Xanthobacter sp. C20 contains a novel bioconversion pathway for limonene. J. Biotechnol. 2000, 84, 133–143. [Google Scholar] [CrossRef]
- Molina, G.; Pessôa, M.G.; Bicas, J.L.; Fontanille, P.; Larroche, C.; Pastore, G.M. Optimization of limonene biotransformation for the production of bulk amounts of α-terpineol. Bioresour. Technol. 2019, 294, 122180. [Google Scholar] [CrossRef]
- Yang, E.-J.; Park, Y.-J.; Chang, H.-C. Cloning of four genes involved in limonene hydroxylation from Enterobacter cowanii 6L. J. Microbiol. Biotechnol. 2007, 17, 1169–1176. [Google Scholar]
- Van der Werf, M.J.; Boot, A.M. Metabolism of carveol and dihydrocarveol in Rhodococcus erythropolis DCL14. Microbiology 2000, 146, 1129–1141. [Google Scholar] [CrossRef] [Green Version]
- Noma, Y. Conversion of (−)-carvone by strains of Streptomyces, A-5–1, and Nocardia, 1–3–11. Agric. Biol. Chem. 1980, 44, 807–812. [Google Scholar] [CrossRef]
- Van der Vlugt-Bergmans, C.J.B.; van der Werf, M.J. Genetic and biochemical characterization of a novel monoterpene epsilon-lactone hydrolase from Rhodococcus erythropolis DCL14. Appl. Environ. Microbiol. 2001, 67, 733–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morrish, J.L.E.; Brennan, E.T.; Dry, H.C.; Daugulis, A.J. Enhanced bioproduction of carvone in a two-liquid-phase partitioning bioreactor with a highly hydrophobic biocatalyst. Biotechnol. Bioeng. 2008, 101, 768–775. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.C.; Da Fonseca, C.O.; Schönthal, A.H. Preclinical development and clinical use of perillyl alcohol for chemoprevention and cancer therapy. Am. J. Cancer Res. 2015, 5, 1580–1593. [Google Scholar] [PubMed]
- Van Beilen, J.B.; Holtackers, R.; Lüscher, D.; Bauer, U.; Witholt, B.; Duetz, W.A. Biocatalytic production of perillyl alcohol from limonene by using a novel Mycobacterium sp. cytochrome P450 alkane hydroxylase expressed in Pseudomonas putida. Appl. Environ. Microbiol. 2005, 71, 1737–1744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornelissen, S.; Julsing, M.K.; Volmer, J.; Riechert, O.; Schmid, A.; Bühler, B. Whole-cell-based CYP153A6-catalyzed (S)-limonene hydroxylation efficiency depends on host background and profits from monoterpene uptake via AlkL. Biotechnol. Bioeng. 2013, 110, 1282–1292. [Google Scholar] [CrossRef]
- Speelmans, G.; Bijlsma, A.; Eggink, G. Limonene bioconversion to high concentrations of a single and stable product, perillic acid, by a solvent-resistant Pseudomonas putida strain. Appl. Microbiol. Biotechnol. 1998, 50, 538–544. [Google Scholar] [CrossRef]
- Monteiro, J.L.F.; Veloso, C.O. Catalytic conversion of terpenes into fine chemicals. Top. Catal. 2004, 27, 169–180. [Google Scholar] [CrossRef]
- Bicas, J.L.; Fontanille, P.; Pastore, G.M.; Larroche, C. A bioprocess for the production of high concentrations of R-(+)-α-terpineol from R-(+)-limonene. Process. Biochem. 2010, 45, 481–486. [Google Scholar] [CrossRef]
- Peterson, J.A.; Lu, J.-Y.; Geisselsoder, J.; Graham-Lorence, S.; Carmona, C.; Witney, F.; Lorence, M.C. Cytochrome P-450terp. Isolation and purification of the protein and cloning and sequencing of its operon. J. Biol. Chem. 1992, 267, 14193–14203. [Google Scholar] [PubMed]
- Hasemann, C.A.; Ravichandran, K.G.; Peterson, J.A.; Deisenhofer, J. Crystal structure and refinement of cytochrome P450terp at 2·3 Å resolution. J. Mol. Biol. 1994, 236, 1169–1185. [Google Scholar] [CrossRef]
- Narushima, H.; Omori, T.; Minoda, Y. Microbial oxidation of beta-myrcene. In Advances in Biotechnology; Vezina, C., Singh, K., Eds.; Pergamon Press: Oxford, UK, 1982; pp. 525–531. [Google Scholar] [CrossRef]
- Iurescia, S.; Marconi, A.M.; Tofani, D.; Gambacorta, A.; Paternò, A.; Devirgiliis, C.; van der Werf, M.J.; Zennaro, E. Identification and sequencing of β-myrcene catabolism genes from Pseudomonas sp. strain M1. Appl. Environ. Microbiol. 1999, 65, 2871–2876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esmaeili, A.; Hashemi, E. Biotransformation of myrcene by Pseudomonas aeruginosa. Chem. Cent. J. 2011, 5, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, M.L.; Marriott, R.; Dowle, A.; Grogan, G. Biotransformation of β-myrcene to geraniol by a strain of Rhodococcus erythropolis isolated by selective enrichment from hop plants. Appl. Microbiol. Biotechnol. 2010, 85, 721–730. [Google Scholar] [CrossRef]
- Marmulla, R.; Šafarić, B.; Markert, S.; Schweder, T.; Harder, J. Linalool isomerase, a membrane-anchored enzyme in the anaerobic monoterpene degradation in Thauera linaloolentis 47Lol. BMC Biochem. 2016, 17, 6. [Google Scholar] [CrossRef] [Green Version]
- Bell, S.G.; Dale, A.; Rees, N.H.; Wong, L.-L. A cytochrome P450 class I electron transfer system from Novosphingobium aromaticivorans. Appl. Microbiol. Biotechnol. 2010, 86, 163–175. [Google Scholar] [CrossRef]
- Ullah, A.J.; Murray, R.I.; Bhattacharyya, P.K.; Wagner, G.C.; Gunsalus, I.C. Protein components of a cytochrome P-450 linalool 8-methyl hydroxylase. J. Biol. Chem. 1990, 265, 1345–1351. [Google Scholar]
- Lüddeke, F.; Wülfing, A.; Timke, M.; Germer, F.; Weber, J.; Dikfidan, A.; Rahnfeld, T.; Linder, D.; Meyerdierks, A.; Harder, J. Geraniol and geranial dehydrogenases induced in anaerobic monoterpene degradation by Castellaniella defragrans. Appl. Environ. Microbiol. 2012, 78, 2128–2136. [Google Scholar] [CrossRef] [Green Version]
- Renganathan, V.; Madyastha, K.M. Linalyl acetate is metabolized by Pseudomonas incognita with the acetoxy group intact. Appl. Environ. Microbiol. 1983, 45, 6–15. [Google Scholar] [CrossRef] [Green Version]
- Förster-Fromme, K.; Höschle, B.; Mack, C.; Bott, M.; Armbruster, W.; Jendrossek, D. Identification of genes and proteins necessary for catabolism of acyclic terpenes and leucine/isovalerate in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2006, 72, 4819–4828. [Google Scholar] [CrossRef] [Green Version]
- Joglekar, S.S.; Dhavlikar, R.S. Microbial transformation of terpenoids. I. Indentification of metabolites produced by a pseudomonad from citronellal and citral. Appl. Microbiol. 1969, 18, 1084–1087. [Google Scholar] [CrossRef] [PubMed]
- Santos, P.M.; Sá-Correia, I. Adaptation to β-myrcene catabolism in Pseudomonas sp. M1: An expression proteomics analysis. Proteomics 2009, 9, 5101–5111. [Google Scholar] [CrossRef]
- Soares-Castro, P.; Santos, P.M. Deciphering the genome repertoire of Pseudomonas sp. M1 toward β-myrcene biotransformation. Genome Biol. Evol. 2015, 7, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Ivarsson, P.; Birgersson, G. Regulation and biosynthesis of pheromone components in the double spined bark beetle Ips duplicatus (Coleoptera: Scolytidae). J. Insect Physiol. 1995, 41, 843–849. [Google Scholar] [CrossRef]
- Kafka, W.A.; Ohloff, G.; Schneider, D.; Vareschi, E. Olfactory discrimination of two enantiomers of 4-methyl-hexanoic acid by the migratory locust and the honeybee. J. Comp. Physiol. 1973, 87, 277–284. [Google Scholar] [CrossRef]
- Miyazato, H.; Hashimoto, S.; Hayashi, S. First identification of the odour-active unsaturated aliphatic acid (E)-4-methyl-3-hexenoic acid in yuzu (Citrus junos Sieb. ex Tanaka). Flavour Fragr. J. 2013, 28, 62–69. [Google Scholar] [CrossRef]
- Lüddeke, F.; Harder, J. Enantiospecific (S)-(+)-linalool formation from beta-myrcene by linalool dehydratase-isomerase. Z Naturforsch C 2011, 66, 409–412. [Google Scholar] [CrossRef]
- Sanchez, S.; Demain, A.L. Enzymes and bioconversions of industrial, pharmaceutical, and biotechnological significance. Org. Process. Res. Dev. 2011, 15, 224–230. [Google Scholar] [CrossRef]
- Dudley, Q.M.; Karim, A.S.; Jewett, M.C. Cell-free metabolic engineering: Biomanufacturing beyond the cell. Biotechnol. J. 2015, 10, 69–82. [Google Scholar] [CrossRef]
- Johannes, T.W.; Zhao, H. Directed evolution of enzymes and biosynthetic pathways. Curr. Opin. Microbiol. 2006, 9, 261–267. [Google Scholar] [CrossRef]
- Böttcher, D.; Bornscheuer, U.T. Protein engineering of microbial enzymes. Curr. Opin. Microbiol. 2010, 13, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Woolston, B.M.; Edgar, S.; Stephanopoulos, G. Metabolic engineering: Past and future. Annu. Rev. Chem. Biomol. Eng. 2013, 4, 259–288. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhou, Y.J.; Siewers, V.; Nielsen, J. Enabling technologies to advance microbial isoprenoid production. In Biotechnology of Isoprenoids; Springer: Cham, Switzerland, 2014; pp. 143–160. [Google Scholar] [CrossRef]
- Janocha, S.; Schmitz, D.; Bernhardt, R. Terpene hydroxylation with microbial cytochrome P450 monooxygenases. In Biotechnology of Isoprenoids; Springer: Cham, Switzerland, 2015; pp. 215–250. [Google Scholar] [CrossRef]
- Hernandez-Ortega, A.; Vinaixa, M.; Zebec, Z.; Takano, E.; Scrutton, N.S. A toolbox for diverse oxyfunctionalisation of monoterpenes. Sci. Rep. 2018, 8, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bell, S.G.; Chen, X.; Sowden, R.J.; Xu, F.; Williams, J.N.; Wong, L.-L.; Rao, Z. Molecular recognition in (+)-α-pinene oxidation by cytochrome P450cam. J. Am. Chem. Soc. 2003, 125, 705–714. [Google Scholar] [CrossRef]
- Harford-Cross, C.F.; Carmichael, A.B.; Allan, F.K.; England, P.A.; Rouch, D.A.; Wong, L.-L. Protein engineering of cytochrome P450cam (CYP101) for the oxidation of polycyclic aromatic hydrocarbons. Protein Eng. 2000, 13, 121–128. [Google Scholar] [CrossRef] [Green Version]
- Jones, J.P.; O’Hare, E.J.; Wong, L. Oxidation of polychlorinated benzenes by genetically engineered CYP101 (cytochrome P450cam). Eur. J. Biochem. 2001, 268, 1460–1467. [Google Scholar] [CrossRef]
- Xu, F.; Bell, S.G.; Lednik, J.; Insley, A.; Rao, Z.; Wong, L. The heme monooxygenase cytochrome P450cam can be engineered to oxidize ethane to ethanol. Angew. Chem. 2005, 44, 4029–4032. [Google Scholar] [CrossRef]
- Whitehouse, C.J.C.; Bell, S.G.; Wong, L.-L. P450 BM3 (CYP102A1): Connecting the dots. Chem. Soc. Rev. 2012, 41, 1218–1260. [Google Scholar] [CrossRef]
- Watanabe, Y.; Laschat, S.; Budde, M.; Affolter, O.; Shimada, Y.; Urlacher, V.B. Oxidation of acyclic monoterpenes by P450 BM-3 monooxygenase: Influence of the substrate E/Z-isomerism on enzyme chemo-and regioselectivity. Tetrahedron 2007, 63, 9413–9422. [Google Scholar] [CrossRef]
- Belsare, K.D.; Horn, T.; Ruff, A.J.; Martinez, R.; Magnusson, A.; Holtmann, D.; Schrader, J.; Schwaneberg, U. Directed evolution of P450cin for mediated electron transfer. Protein Eng. Des. Sel. 2017, 30, 119–127. [Google Scholar] [CrossRef] [Green Version]
- Arand, M.; Hallberg, B.M.; Zou, J.; Bergfors, T.; Oesch, F.; van der Werf, M.J.; de Bont, J.A.M.; Jones, T.A.; Mowbray, S.L. Structure of Rhodococcus erythropolis limonene-1, 2-epoxide hydrolase reveals a novel active site. EMBO J. 2003, 22, 2583–2592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, H.; Reetz, M.T. Manipulating the stereoselectivity of limonene epoxide hydrolase by directed evolution based on iterative saturation mutagenesis. J. Am. Chem. Soc. 2010, 132, 15744–15751. [Google Scholar] [CrossRef] [PubMed]
- Weidenweber, S.; Marmulla, R.; Ermler, U.; Harder, J. X-ray structure of linalool dehydratase/isomerase from Castellaniella defragrans reveals enzymatic alkene synthesis. Febs Lett. 2016, 590, 1375–1383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soares-Castro, P. Optimization of Pseudomonas sp. M1 as a Biocatalyst of β-Myrcene and Related Plantderived Volatiles Using Synthetic Biology. Ph.D. Thesis, University of Minho, Braga, Portugal, 2017. [Google Scholar]
- Siedenburg, G.; Jendrossek, D.; Breuer, M.; Juhl, B.; Pleiss, J.; Seitz, M.; Klebensberger, J.; Hauer, B. Activation-independent cyclization of monoterpenoids. Appl. Environ. Microbiol. 2012, 78, 1055–1062. [Google Scholar] [CrossRef] [Green Version]
- Bastian, S.A.; Hammer, S.C.; Kreß, N.; Nestl, B.M.; Hauer, B. Selectivity in the cyclization of citronellal introduced by squalene hopene cyclase variants. ChemCatChem 2017, 9, 4364–4368. [Google Scholar] [CrossRef]
- Uchiyama, T.; Miyazaki, K. Functional metagenomics for enzyme discovery: Challenges to efficient screening. Curr. Opin. Biotechnol. 2009, 20, 616–622. [Google Scholar] [CrossRef]
- Cheng, X.-Y.; Tian, X.-L.; Wang, Y.-S.; Lin, R.-M.; Mao, Z.-C.; Chen, N.; Xie, B.-Y. Metagenomic analysis of the pinewood nematode microbiome reveals a symbiotic relationship critical for xenobiotics degradation. Sci. Rep. 2013, 3, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Adams, A.S.; Boone, C.K.; Bohlmann, J.; Raffa, K.F. Responses of bark beetle-associated bacteria to host monoterpenes and their relationship to insect life histories. J. Chem. Ecol. 2011, 37, 808–817. [Google Scholar] [CrossRef]
- Vilanova, C.; Marín, M.; Baixeras, J.; Latorre, A.; Porcar, M. Selecting microbial strains from pine tree resin: Biotechnological applications from a terpene world. PLoS ONE 2014, 9, e100740. [Google Scholar] [CrossRef] [Green Version]
- Calero, P.; Nikel, P.I. Chasing bacterial chassis for metabolic engineering: A perspective review from classical to non-traditional microorganisms. Microb. Biotechnol. 2019, 12, 98–124. [Google Scholar] [CrossRef]
- Kampers, L.F.C.; Volkers, R.J.M.; Martins dos Santos, V.A.P. Pseudomonas putida KT2440 is HV 1 certified, not GRAS. Microb. Biotechnol. 2019, 12, 845–848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hick, A.J.; Luszniak, M.C.; Pickett, J.A. Volatile isoprenoids that control insect behaviour and development. Nat. Prod. Rep. 1999, 16, 39–54. [Google Scholar] [CrossRef]
- Mori, K.; Igarashi, Y. Pheromone synthesis, CIV. Synthesis of the enantiomers of α-phellandren-8-ol (p-mentha-1,5-dien-8-ol), a monoterpene from bark beetles. Liebigs Ann. der Chem. 1988, 1988, 93–95. [Google Scholar] [CrossRef]
- Van Hecke, W.; Kaur, G.; De Wever, H. Advances in in-situ product recovery (ISPR) in whole cell biotechnology during the last decade. Biotechnol. Adv. 2014, 32, 1245–1255. [Google Scholar] [CrossRef] [PubMed]
- De Carvalho, C.C.R.; Van Keulen, F.; Manuela, M.; da Fonseca, R. Production and recovery of limonene-1, 2-diol and simultaneous resolution of a diastereomeric mixture of limonene-1, 2-epoxide with whole cells of Rhodococcus erythropolis DCL14. Biocatal. Biotransform. 2000, 18, 223–235. [Google Scholar] [CrossRef]
- Muntendam, R.; Melillo, E.; Ryden, A.; Kayser, O. Perspectives and limits of engineering the isoprenoid metabolism in heterologous hosts. Appl. Microbiol. Biotechnol. 2009, 84, 1003. [Google Scholar] [CrossRef]
- Korman, T.P.; Opgenorth, P.H.; Bowie, J.U. A synthetic biochemistry platform for cell free production of monoterpenes from glucose. Nat. Commun. 2017, 8, 1–8. [Google Scholar] [CrossRef]
- Schwab, W.; Davidovich-Rikanati, R.; Lewinsohn, E. Biosynthesis of plant-derived flavor compounds. Plant J. 2008, 54, 712–732. [Google Scholar] [CrossRef]
- Yang, J.; Nie, Q.; Ren, M.; Feng, H.; Jiang, X.; Zheng, Y.; Liu, M.; Zhang, H.; Xian, M. Metabolic engineering of Escherichia coli for the biosynthesis of alpha-pinene. Biotechnol. Biofuels 2013, 6, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Niu, F.-X.; He, X.; Wu, Y.-Q.; Liu, J.-Z. Enhancing production of pinene in Escherichia coli by using a combination of tolerance, evolution, and modular co-culture engineering. Front. Microbiol. 2018, 9, 1623. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, Q.; Cao, Y.; Feng, X.; Zheng, Y.; Zou, H.; Liu, H.; Yang, J.; Xian, M. Microbial production of sabinene—A new terpene-based precursor of advanced biofuel. Microb. Cell Fact. 2014, 13, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willrodt, C.; David, C.; Cornelissen, S.; Bühler, B.; Julsing, M.K.; Schmid, A. Engineering the productivity of recombinant Escherichia coli for limonene formation from glycerol in minimal media. Biotechnol. J. 2014, 9, 1000–1012. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.-M.; Eom, J.-H.; Um, Y.; Kim, Y.; Woo, H.M. Microbial synthesis of myrcene by metabolically engineered Escherichia coli. J. Agric. Food Chem. 2015, 63, 4606–4612. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, C.; Yoon, S.-H.; Jang, H.-J.; Choi, E.-S.; Kim, S.-W. Engineering Escherichia coli for selective geraniol production with minimized endogenous dehydrogenation. J. Biotechnol. 2014, 169, 42–50. [Google Scholar] [CrossRef]
- Mi, J.; Becher, D.; Lubuta, P.; Dany, S.; Tusch, K.; Schewe, H.; Buchhaupt, M.; Schrader, J. De novo production of the monoterpenoid geranic acid by metabolically engineered Pseudomonas putida. Microb. Cell Fact. 2014, 13, 170. [Google Scholar] [CrossRef] [Green Version]
- Davies, F.K.; Work, V.H.; Beliaev, A.S.; Posewitz, M.C. Engineering limonene and bisabolene production in wild type and a glycogen-deficient mutant of Synechococcus sp. PCC 7002. Front. Bioeng. Biotechnol. 2014, 2, 21. [Google Scholar] [CrossRef] [Green Version]
- Alonso-Gutierrez, J.; Chan, R.; Batth, T.S.; Adams, P.D.; Keasling, J.D.; Petzold, C.J.; Lee, T.S. Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metab. Eng. 2013, 19, 33–41. [Google Scholar] [CrossRef]







| Compound | Aroma | Observations of Biotech Production |
|---|---|---|
| Phenolic aldehydes | ||
| vanillin | vanilla aroma | biotech production established by, e.g., Evolva-IFF, Solvay, Mane, Shangai Apple, BASF, Isobionics |
| safranal | saffron aroma, sweet, spicy, floral odor with a bitter taste | biotech production announced by, e.g., Evolva |
| Lactones | ||
| γ-decalactone | fruity, peach-like aroma | biotech production established by, e.g., BASF, Symrise |
| γ-undecalactone | fruity, sweat peach-like aroma | - |
| Ketones | ||
| 2-heptanone | fruity, cinnamon, banana-like | - |
| α- and β-Ionone | woody, raspberry-type, floral, violet-like odor | - |
| nootkatone | citrusy notes and grapefruit-like aroma | biotech production established by, e.g., Allylix (Evolva), Isobionics, Oxford Biotrans |
| Alcohols | ||
| 1-octen-3-ol | sweet, earthy, herbaceous floral notes, reminiscent of lavender | - |
| Carboxylic acids | ||
| citric acid | acid taste; odorless | - |
| Esters | ||
| ethyl butanoate | sweet and pineapple-like aroma | - |
| Essential Oils | ||
| orange peel oil | orange aroma | - |
| lemon peel oil | lemon aroma | - |
| eucalyptus oil | camphoraceous odor, spicy, cooling taste | - |
| peppermint oil | odor of peppermint, cooling, minty, menthol, sweet taste | - |
| spearmint oil | minty, carvone-like, cooling, candy, spicy | - |
| Monoterpenes | ||
| α-pinene | terpy, citrus and spicy, woody pine and turpentine-like with a slight cooling camphoraceous nutmeg-like note | - |
| β-pinene | cooling, woody, piney and turpentine-like with a fresh minty, eucalyptus and camphoraceous note | - |
| 1,8-cineole | cooling, fresh, oily, green, spicy, pine-like | - |
| limonene | (+)-limonene has an orange-like odor (−)-limonene has a more harsh turpentine-like odor with a lemon note | - |
| (−)-menthol | minty, coolant odor | - |
| menthone | minty, cooling, sweet, peppermint, camphoraceous aroma with a green herbal anise nuance | - |
| carvone | (R)-(−)-carvone has a spearmint aroma (S)-(+)-carvone has a caraway aroma | - |
| α-terpineol | pine odor, floral aroma | - |
| β-myrcene | terpy, herbaceous, woody odor with a mango-like nuance. | - |
| linalool | floral, fresh, sweet, citrus-like aroma | - |
| citronellol | rose-like scent | - |
| citral | lemon, peely, citrus, floral with woody and candy notes. | - |
| geraniol | rose-like, sweet, fruity aroma | - |
| Sesquiterpenes | ||
| α-farnesene | dry woody, green leafy, herbal and floral nuance | biotech production established by, e.g., Amyris-Antibióticos S.A |
| (+)-valencene | sweet, fresh, grapefruit-like aroma | biotech production established by, e.g., Allylix (Evolva), Isobionics |
| Whole-Cell Biocatalyst | Substrate | Product | Ref. |
|---|---|---|---|
| Pseudomonas sp. TK2102 | Eugenol | vanillin | [42] |
| (JP patent 5227980) | |||
| Pseudomonas putida ATCC55180 | Eugenol | vanillin | [43] |
| (US patent 5128253) | Ferulic acid | ||
| Pseudomonas sp. NCIB 11671 | α- and β-Pinene | (−)-carvone (spearmint aroma) | [44] |
| (US patent 4495284) | |||
| Enzymatic Biocatalyst | Substrate | Product | Ref. |
| Commercial lipase AK from Pseudomonas fluorescens (Amano Enzyme Inc.) | (±)-menthol, | (−)-menthyl acetate for the production of (−)-menthol | [45] |
| (±)-neomenthol, | |||
| (±)-neoisomenthol, | |||
| (±)-isomenthol | |||
| Commercial lipase PS from Burkholderia cepacia (Amano Enzyme Inc.) | (±)-isopulegol isomers and vinyl acetate | (−)-isopulegol acetate for the production of (−)-isopulegol | [46] |
| Commercial lipase PS from Burkholderia cepacia (Amano Enzyme Inc.) | (±)-mentholand vinyl acetate | (+)-menthol and(−)-menthyl acetate for the production of (−)-menthol | [46] |
| Commercial esterase from Bacillus subtilis ECU0554 | (±)-menthol esters | (−)-menthol | [47] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soares-Castro, P.; Soares, F.; Santos, P.M. Current Advances in the Bacterial Toolbox for the Biotechnological Production of Monoterpene-Based Aroma Compounds. Molecules 2021, 26, 91. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules26010091
Soares-Castro P, Soares F, Santos PM. Current Advances in the Bacterial Toolbox for the Biotechnological Production of Monoterpene-Based Aroma Compounds. Molecules. 2021; 26(1):91. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules26010091
Chicago/Turabian StyleSoares-Castro, Pedro, Filipa Soares, and Pedro M. Santos. 2021. "Current Advances in the Bacterial Toolbox for the Biotechnological Production of Monoterpene-Based Aroma Compounds" Molecules 26, no. 1: 91. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules26010091