Evaluation of Antiproliferative Palladium(II) Complexes of Synthetic Bisdemethoxycurcumin towards In Vitro Cytotoxicity and Molecular Docking on DNA Sequence
Abstract
:1. Introduction
2. Results and Discussion
2.1. Preparation and Structural Characterization of Palladium(II) Complexes
2.2. DFT Calculations
2.3. Docking Calculations
2.3.1. Human Serum Albumin (HSA)
2.3.2. Transcription Factor NF-κB
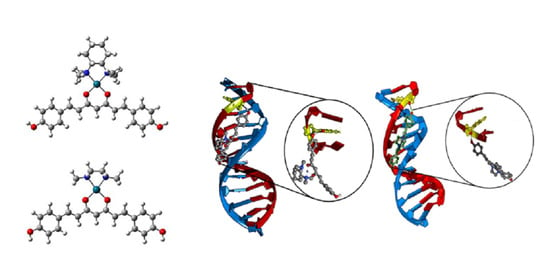
2.3.3. DNA Sequence
2.4. Biological Tests
3. Materials and Methods
3.1. Synthesis of Palladium(II) Complexes
3.2. X-ray Crystallography
3.3. Computational Details
3.4. Biological Testing
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Lippert, B. Cisplatin, Chemistry and Biochemistry of a Leading Anticancer Drug; Wiley-VCH: Weinheim, Germany, 1991. [Google Scholar]
- Todd, R.C.; Lippard, S.J. Inhibition of transcription by platinum antitumor compounds. Metallomics 2009, 1, 280–291. [Google Scholar] [CrossRef] [Green Version]
- Florea, A.-M.; Büsselberg, D. Cisplatin as an anti-tumordrug: Cellular mechanisms of activity, drug resistance and induced side effects. Cancer 2011, 3, 1351–1371. [Google Scholar] [CrossRef] [PubMed]
- Monneret, C. Platinum anticancer drugs. From serendipity to rational design. Ann. Pharm. Fr. 2011, 69, 286–295. [Google Scholar] [CrossRef]
- Chan, B.A.; Coward, J.I.G. Review article chemotherapy advances in small-cell lung cancer. J. Thorac. Dis. 2007, 5, S565–S578. [Google Scholar]
- Uehara, T.; Yamate, J.; Torii, M.; Maruyama, T. Comparative nephrotoxicity of cisplatin and nedaplatin: Mechanisms and histopathological characteristics. J. Toxicol. Pathol. 2011, 24, 87–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnstone, T.C.; Suntharalingam, K.; Lippard, S.J. The next generation of platinum drugs: Targeted Pt(II) agents, nanoparticle delivery, and Pt(IV) prodrugs. Chem. Rev. 2016, 116, 3436–3486. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; Liu, Y.E.; Ren, X.C.; Chen, X.J.; Su, H.L.; Zong, J.; Feng, Z.L.; Wang, D.Y.; Lin, Q.; Gao, X.S. A phase I clinical trial of dose escalation of loboplatin in combination with fixed—Dose docetaxel for the treatment of human solid tumors that had progressed following chemotherapy. Oncol. Lett. 2015, 9, 67–74. [Google Scholar] [CrossRef] [Green Version]
- Espina, M.; Corte-Rodríguez, M.; Aguado, L.; Montes-Bayón, M.; Sierra, M.I.; Martínez-Camblor, P.; Blanco-González, E.; Sierra, L.M. Cisplatin resistance in cell models: Evaluation of metallomic and biological predictive biomarkers to address early therapy failure. Metallomics 2017, 9, 564–574. [Google Scholar] [CrossRef] [Green Version]
- Moulick, A.; Heger, Z.; Milosavljevic, V.; Richtera, L.; Barroso-Flores, J.; Merlos Rodrigo, M.A.; Buchtelova, H.; Podgajny, R.; Hynek, D.; Kopel, P.; et al. Real-Time Visualization of Cell Membrane Damage Using Gadolinium–Schiff Base Complex-Doped Quantum Dots. ACS Appl. Mater. Interfaces 2018, 10, 35859–35868. [Google Scholar] [CrossRef]
- Li, S.; Chen, Y.; Zhu, W.; Yang, W.; Chen, Z.; Song, J.; Song, X.; Chen, X.; Yang, H. Engineered Nanoscale Vanadium Metallodrugs for Robust Tumor-Specific Imaging and Therapy. Adv. Funct. Mater. 2021, 31, 2010337. [Google Scholar] [CrossRef]
- Martínez-Valencia, B.; Corona-Motolinia, N.D.; Sánchez-Lara, E.; Noriegab, L.; Sánchez-Gaytán, B.L.; Castro, M.E.; Meléndez-Bustamante, F.; González-Vergara, E. Cyclo-tetravanadate bridged copper complexes as potential double bullet pro-metallodrugs for cancer treatment. J. Inorg. Biochem. 2020, 208, 111081. [Google Scholar] [CrossRef] [PubMed]
- Allardyce, C.S.; Dyson, P.J. Metal-based drugs that break the rules. Dalton Trans. 2016, 45, 3201–3209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Xu, C.; Li, T.; Lu, S.; Luo, F.; Wang, H. Novel NHC-coordinated ruthenium(II) arene complexes achieve synergistic efficacy as safe and effective anticancer therapeutics. Eur. J. Med. Chem. 2020, 203, 112605. [Google Scholar] [CrossRef] [PubMed]
- Morais, T.S.; Valente, A.; Tomaz, A.I.; Marques, F.; Garcia, M.H. Tracking antitumor metallodrugs: Promising agents with the Ru(II)- and Fe(II)-cyclopentadienyl scaffolds. Future Med. Chem. 2016, 8, 527–544. [Google Scholar] [CrossRef]
- Murray, B.S.; Crot, S.; Siankevich, S.; Dyson, P.J. Potential of cycloaddition reactions to generate cytotoxic metal drugs in vitro. Inorg. Chem. 2014, 53, 9315–9321. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C.; Karge, B.; Misgeld, R.; Prokop, A.; Franke, R.; Brönstrup, M.; Ott, I. Gold(I) NHC Complexes: Antiproliferative Activity, Cellular Uptake, Inhibition of Mammalian and Bacterial Thioredoxin Reductases, and Gram-Positive Directed Antibacterial Effects. Chem. A Eur. J. 2017, 23, 1869–1880. [Google Scholar] [CrossRef]
- Nobili, S.; Mini, E.; Landini, I.; Gabbiani, C.; Casini, A.; Messori, L. Gold compounds as anticancer agents: Chemistry, cellular pharmacology, and preclinical studies. Med. Res. Rev. 2010, 30, 550–580. [Google Scholar] [CrossRef]
- Zou, T.; Ching, A.; Lum, T.; Lok, C.-N.; Zhang, J.-J.; Che, C.-M. Chemical biology of anticancer gold(III) and gold(I) complexes. Chem. Soc. Rev. 2015, 44, 8786–8801. [Google Scholar] [CrossRef] [PubMed]
- Kalinowska-Lis, U.; Felczak, A.; Checinska, L.; Szabłowska-Gadomska, I.; Patyna, E.; Małecki, M.; Lisowska, K.; Ochocki, J. Antibacterial activity and cytotoxicity of silver(I) complexes of pyridine and (Benz)imidazole derivatives. X-ray crystal structure of [Ag(2,6-di(CH(2)OH)py)(2)]NO(3). Molecules 2015, 21, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Starha, P.; Travnicek, Z. Non-platinum complexes containing releasable biologically active ligands. Coord. Chem. Rev. 2019, 395, 130–145. [Google Scholar] [CrossRef]
- Vojtek, M.; Marques, M.P.M.; Ferreira, I.M.P.L.V.O.; Mota-Filipe, H.; Diniz, C. Anticancer activity of palladium-based complexes against triple-negative breast cancer. Drug Discov. Today 2019, 24, 1044–1058. [Google Scholar] [CrossRef]
- Malinowska, K.; Szczepanska, A.; Zielinska-Blizniewska, H.; Majsterek, I. An Evaluation of the Antioxidant and Anticancer Properties of Complex Compounds of Copper (II), Platinum (II), Palladium (II) and Ruthenium (III) for Use in Cancer Therapy. Mini Rev. Med. Chem. 2018, 18, 1373–1381. [Google Scholar]
- Liu, Y.; Li, J.; Chen, M.; Chen, X.; Zheng, N. Palladium-based nanomaterials for cancer imaging and therapy. Theranostics 2020, 10, 10057–10074. [Google Scholar] [CrossRef]
- Yaqoob, S.B.; Adnan, R.; Khan, R.M.R.; Rashid, M. Gold, Silver, and Palladium Nanoparticles: A Chemical Tool for Biomedical Applications. Front. Chem. 2020, 8, 376. [Google Scholar] [CrossRef] [PubMed]
- Dupont, J.; Consorti, C.S.; Spencer, J. The potential of palladacycles: More than just precatalysts. Chem. Rev. 2005, 105, 2527–2572. [Google Scholar] [CrossRef]
- Pucci, D.; Bloise, R.; Bellusci, A.; Bernardini, S.; Ghedini, M.; Pirillo, S.; Valentini, A.; Crispini, A. Curcumin and cyclopalladated complexes: A recipe for bifunctional biomaterials. J. Inorg. Biochem. 2007, 101, 1013–1022. [Google Scholar] [CrossRef]
- Valentini, A.; Conforti, F.; Crispini, A.; De Martino, A.; Condello, R.; Stelliano, C.; Rotilio, G.; Ghedini, M.; Federici, G.; Bernardini, S.; et al. Synthesis, Oxidant Properties, and Antitumoral Effects of a Heteroleptic Palladium(II) Complex of Curcumin on Human Prostate Cancer Cells. J. Med. Chem. 2009, 52, 484–491. [Google Scholar] [CrossRef]
- Miklášová, N.; Mikláš, R.; Devínsky, F. Palladium Complexes of Curcumin and Its Analogues and Methods of Preparation of the Same. WO 2014/175841A1, 30 October 2014. [Google Scholar]
- Miklášová, N.; Fischer-Fodor, E.; Mikláš, R.; Kucková, L.; Kožíšek, J.; Liptaj, T.; Soritau, O.; Valentová, J.; Devínsky, F. Synthesis and characterization of new biologically active palladium(II) complexes with (1E,6E)-1,7-bis(3,4-diethoxyphenyl)-1,6-heptadiene-3,5-dione. Inorg. Chem. Commun. 2014, 46, 229–233. [Google Scholar] [CrossRef]
- Fischer-Fodor, E.; Mikláš, R.; Krausz, L.T.; Virag, P.; Moldovan, D.C.; Perde Schrepler, M.; Berindan-Neagoe, I.; Devínsky, F.; Miklášová, N. Immunomodulatory potential of Palladium(II) complexes with (1E,6E)-1,7-bis(3,4-dimethoxyphenyl)hepta-1,6-diene-3,5-dione. Stud. Univ. Babeş-Bolyai Chem. 2015, 60, 93–100. [Google Scholar]
- Miklášová, N.; Mikláš, R.; Virag, P.; Tatomir, C.B.; Szalontai, C.; Cenariu, D.; Devínsky, F.; Fischer-Fodor, E. Cytotoxic activity of palladium(II) complexes of (1E,6E)-1,7-bis(4-(dimethylamino)phenyl)hepta-1,6-diene-3,5-dione against human colon carcinoma. Stud. Univ. Babeş-Bolyai Chem. 2016, 61, 109–116. [Google Scholar]
- Fischer-Fodor, E.; Mikláš, R.; Rišiaňová, L.; Cenariu, M.; Grosu, I.-G.; Virag, P.; Perde Schrepler, M.; Tomuleasa, C.; Berindan-Neagoe, I.; Devínsky, F.; et al. Novel palladium(II) complexes that influence prominin-1/CD133 expression and stem cell factor release in tumor cells. Molecules 2017, 22, 561. [Google Scholar] [CrossRef]
- Ramezani, M.; Hatamipour, M.; Sahebkar, A. Promising Anti-Tumor Properties of Bisdemethoxycurcumin: A Naturally Occurring Curcumin Analogue. J. Cell. Physiol. 2018, 233, 880–887. [Google Scholar] [CrossRef]
- Qiu, C.; Liu, K.; Zhang, S.; Gao, S.; Chen, W.; Li, D.; Huang, Y. Bisdemethoxycurcumin Inhibits Hepatocellular Carcinoma Proliferation Through Akt Inactivation via CYLD-Mediated Deubiquitination. Drug Des. Dev. Ther. 2020, 14, 993–1001. [Google Scholar] [CrossRef] [Green Version]
- Liao, C.L.; Chu, Y.L.; Lin, H.Y.; Chen, C.Y.; Hsu, M.J.; Liu, K.C.; Lai, K.C.; Huang, A.C.; Chung, J.G. Bisdemethoxycurcumin Suppresses Migration and Invasion of Human Cervical Cancer Hela Cells via Inhibition of NF-ĸB, MMP-2 and -9 Pathways. Anticancer Res. 2018, 38, 3989–3997. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Yao, L.; Li, J.; Qin, X.; Qiu, Z.; Chen, W. Preparation of Poly(Lactide-Co-Glycolide) Microspheres and Evaluation of Pharmacokinetics and Tissue Distribution of BDMC-PLGA-MS in Rats. Asian J. Pharm. Sci. 2018, 13, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, Q.; Omari-Siaw, E.; Adu-Frimpong, M.; Liu, J.; Xu, X.; Yu, J. Enhanced Oral Bioavailability of Bisdemethoxycurcumin-Loaded Self-Microemulsifying Drug Delivery System: Formulation Design, in Vitro and in Vivo Evaluation. Int. J. Pharm. 2020, 590, 119887. [Google Scholar] [CrossRef] [PubMed]
- Mehanny, M.; Hathout, R.M.; Geneidi, A.S.; Mansour, S. Studying the Effect of Physically-Adsorbed Coating Polymers on the Cytotoxic Activity of Optimized Bisdemethoxycurcumin Loaded-PLGA Nanoparticles. J. Biomed. Mater. Res. Part A 2017, 105, 1433–1445. [Google Scholar] [CrossRef]
- Zhang, Y.; Khan, A.R.; Fu, M.; Zhai, Y.; Yu, A.; Zhai, G. The Progresses in Curcuminoids-Based Metal Complexes: Especially in Cancer Therapy. Future Med. Chem. 2019, 11, 1035–1056. [Google Scholar] [CrossRef]
- Li, Y.; Gu, Z.; Zhang, C.; Li, S.; Zhang, L.; Zhou, G.; Wang, S.; Zhang, J. Synthesis, Characterization and ROS-Mediated Antitumor Effects of Palladium(II) Complexes of Curcuminoids. Eur. J. Med. Chem. 2018, 144, 662–671. [Google Scholar] [CrossRef]
- Al-Ishaq, R.K.; Overy, A.J.; Büsselberg, D. Phytochemicals and Gastrointestinal Cancer: Cellular Mechanisms and Effects to Change Cancer Progression. Biomolecules 2020, 10, 105. [Google Scholar] [CrossRef] [Green Version]
- Liczbiński, P.; Michałowicz, J.; Bukowska, B. Molecular mechanism of curcumin action in signaling pathways: Review of the latest research. Phytother. Res. 2020, 34, 1992–2005. [Google Scholar] [CrossRef] [PubMed]
- Rajagopal, C.; Lankadasari, M.B.; Aranjani, J.M.; Harikumar, K.B. Targeting oncogenic transcription factors by polyphenols: A novel approach for cancer therapy. Pharmacol. Res. 2018, 130, 273–291. [Google Scholar] [CrossRef] [PubMed]
- Latruffe, N.; Menzel, M.; Delmas, D.; Buchet, R.; Lançon, A. Compared binding properties between resveratrol and other polyphenols to plasmatic albumin: Consequences for the health protecting effect of dietary plant microcomponents. Molecules 2014, 19, 17066–17077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irshad, R.; Husain, M. Natural products in the reprogramming of cancer epigenetics. Toxicol. Appl. Pharmacol. 2021, 417, 115467. [Google Scholar] [CrossRef]
- Changtam, C.; de Koning, H.P.; Ibrahim, H.; Sajid, M.S.; Gould, M.K.; Suksamrarn, A. Curcuminoid analogs with potent activity against Trypanosoma and Leishmania species. Eur. J. Med. Chem. 2010, 45, 941–956. [Google Scholar] [CrossRef]
- STOE & Cie GmbH. X-Area 1.84, Software Package for Collecting Single-Crystal Data on STOE Area- Detector Diffractometers, for Image Processing, Scaling Reflection Intensities and for Outlier Rejection; STOE & Cie GmbH: Darmstadt, Germany, 2018. [Google Scholar]
- Dolomanov, H.; Bouhris, O.V.; Gildea, L.J.; Howard, R.J.; Puschmann, J.A.K. OLEX 2. J. Appl. Cryst. 2009, 339–341. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELXT-Integrated space-group and crystal-structure determination. Acta Crystallogr. 2015, 71, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. 2015, 71, 3–8. [Google Scholar]
- Bergerhoff, G.; Berndt, M.; Brandenburg, K. Evaluation of Crystallographic Data with the Program DIAMOND. J. Res. Natl. Inst. Stand. Technol. 1996, 101, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Macrae, C.F.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Shields, G.P.; Taylor, R.; Towler, M.; van de Streek, J. Mercury: Visualization and analysis of crystal structures. J. Appl. Cryst. 2006, 39, 453–457. [Google Scholar] [CrossRef] [Green Version]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16, Revision, C.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Autodock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comp. Chem. 2009, 16, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, P.J.M.; Jamshidi, S.; Farag, D.B. Chapter 2. Computational Approaches in the Development of Small Molecule Small-molecule Transciption Factor Inhibitors. In Drug Discovery Series No 65: Transcription Factor Inhibitors in Oncology; Rahman, K.M., Thurston, D.E., Eds.; The Royal Society of Chemistry: London, UK, 2019; ISBN 978-1-78262-145-4. [Google Scholar]
- Jiang, J.; Pavlova, N.N.; Zhang, J. Asparagine, a critical limiting metabolite during glutamine starvation. Mol. Cell. Oncol. 2018, 5, e1441633. [Google Scholar] [CrossRef] [PubMed] [Green Version]











| Data | Complex 1 | Complex 2 |
|---|---|---|
| Empirical formula | C29H37N2O4Pd | C25H29N2O4Pd |
| Temperature (K)/Wavelength (Å) | 100(1)/1.54186 | 100(1)/1.54186 |
| Crystal system/space group | monoclinic, C2 | monoclinic, P21/n |
| Unit cell dimensions a, b, c (Å) β (°) | a = 26.7722(5) b = 10.2502(1) c = 22.3191(4) β = 100.776(1) | a = 15.4449(1) b = 7.4624(1) c = 25.8668(3) β = 98.978(1) |
| Formula weight/Volume | 584.00/6016.82(2) Å3 | 527.90/2944.78(6) Å3 |
| Z, Calculated density | 8, 1.289 mg/m3 | 4, 1.191 mg/m3 |
| Absorption coefficient/F(000) | 5.238 mm−1/2424 | 5.301 mm−1/1084 |
| Crystal size | 0.26 × 0.1 × 0.07 mm | 0.15 × 0.11 × 0.02 mm |
| 2Θ range for data collection | 6.722 to 143.506° | 6.268 to 143.992° |
| Index ranges | −31 ≤ h ≤ 32 −12 ≤ k ≤ 6 −27 ≤ l ≤ 26 | −19 ≤ h ≤ 13 −9 ≤ k ≤ 8 −22 ≤ l ≤ 31 |
| Reflections collected | 123,856 | 113,553 |
| Independent reflections | 8235[Rint = 0.0388,Rσ = 0.0206] | 5714[Rint = 0.0409,Rσ = 0.0123] |
| Refinement method | Full-matrix least-squares on F2 | Full-matrix least-squares on F2 |
| Data/restraints/parameters | 8235/0/665 | 5714/0/293 |
| Goodness-of-fit on F2 | 1.011 | 1.063 |
| Final R indices [I > 2σ(I)] * | R1 = 0.0376 wR2 = 0.1014 | R1 = 0.0373 wR2 = 0.1225 |
| R indices (all data) | R1 = 0.0459 wR2 = 0.1044 | R1 = 0.0409 wR2 = 0.1277 |
| Flack parameter | 0.024(14) | - |
| Largest diff. peak and hole | 0.72 and −0.70 e. Å−3 | 1.07 and −1.06 e. Å−3 |
| Tested Compounds | |||
|---|---|---|---|
| Cell Lines | 1 | 2 | Cis-Pt |
| A549 | 42.79 ± 10.31 | 86.05 ± 9.85 | 9.50 ± 0.2 |
| Caco-2 | 24.47 ± 5.28 | 78.57 ± 7.63 | 15.20 ± 0.3 |
| HCT | 18.23 ± 4.87 | 38.57 ± 7.84 | 15.30 ± 0.5 |
| HeLa | 17.35 ± 5.54 | 34.81 ± 9.22 | 13.10 ± 0.2 |
| Jurkat | 5.33 ± 0.98 | 7.69 ± 1.56 | 16.20 ± 0.6 |
| MCF-7 | 34.13 ± 7.21 | 94.28 ± 12.87 | 15.60 ± 0.3 |
| MDA | 9.61 ± 1.76 | 50.10 ± 9.67 | 17.50 ± 0.5 |
| 3T3 | 20.32 ± 5.33 | 46.36 ± 8.75 | 20.87 ± 0.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miklášová, N.; Herich, P.; Dávila-Becerril, J.C.; Barroso-Flores, J.; Fischer-Fodor, E.; Valentová, J.; Leskovská, J.; Kožíšek, J.; Takáč, P.; Mojžiš, J. Evaluation of Antiproliferative Palladium(II) Complexes of Synthetic Bisdemethoxycurcumin towards In Vitro Cytotoxicity and Molecular Docking on DNA Sequence. Molecules 2021, 26, 4369. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules26144369
Miklášová N, Herich P, Dávila-Becerril JC, Barroso-Flores J, Fischer-Fodor E, Valentová J, Leskovská J, Kožíšek J, Takáč P, Mojžiš J. Evaluation of Antiproliferative Palladium(II) Complexes of Synthetic Bisdemethoxycurcumin towards In Vitro Cytotoxicity and Molecular Docking on DNA Sequence. Molecules. 2021; 26(14):4369. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules26144369
Chicago/Turabian StyleMiklášová, Natalia, Peter Herich, Juan Carlos Dávila-Becerril, Joaquín Barroso-Flores, Eva Fischer-Fodor, Jindra Valentová, Janka Leskovská, Jozef Kožíšek, Peter Takáč, and Ján Mojžiš. 2021. "Evaluation of Antiproliferative Palladium(II) Complexes of Synthetic Bisdemethoxycurcumin towards In Vitro Cytotoxicity and Molecular Docking on DNA Sequence" Molecules 26, no. 14: 4369. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules26144369