Antioxidant Properties and Aldehyde Reactivity of PD-L1 Targeted Aryl-Pyrazolone Anticancer Agents
Abstract
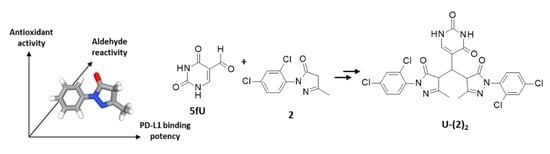
:1. Introduction
2. Results
2.1. Antioxidant Activity
2.1.1. DPPH Assay
2.1.2. DMPO Assay
2.2. Reactivity toward Aromatic Aldehydes
2.2.1. Adduct Formation in the Presence of 5-Formyluracil (5fU) (HRMS Analyses)
2.2.2. Separation and Characterization of Covalent Uracil-Drug Adducts (LC-HRMS Analysis)
2.2.3. Kinetic of Formation of Mono- and Bi-Adducts
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. PD-L1 Binding and PD-L1-Dependent Activity
4.3. EPR Measurements
4.4. Aldehyde Reactivity Measurements
4.4.1. High Resolution Mass Spectrometry (HRMS) and LC-HRMS Methods
4.4.2. HPLC-DAD Method
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yi, M.; Zheng, X.; Niu, M.; Zhu, S.; Ge, H.; Wu, K. Combination strategies with PD-1/PD-L1 blockade: Current advances and future directions. Mol. Cancer 2022, 21, 28. [Google Scholar] [CrossRef] [PubMed]
- Bailly, C.; Thuru, X.; Quesnel, B. Combined cytotoxic chemotherapy and immunotherapy of cancer: Modern times. NAR Cancer 2020, 2, zcaa002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, P.; Khan, F.; Qari, H.A.; Upadhyay, T.K.; Alkhateeb, A.F.; Oves, M. Revolutionization in Cancer Therapeutics via Targeting Major Immune Checkpoints PD-1, PD-L1 and CTLA-4. Pharmaceuticals 2022, 15, 335. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Cheng, Z.; Long, J.; Dömling, A.; Tortorella, M.; Wang, Y. Small Molecule Inhibitors of Programmed Cell Death Ligand 1 (PD-L1): A Patent Review (2019–2021). Expert. Opin. Ther. Pat. 2022, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Skalniak, L.; Zak, K.M.; Guzik, K.; Magiera, K.; Musielak, B.; Pachota, M.; Szelazek, B.; Kocik, J.; Grudnik, P.; Tomala, M.; et al. Small-molecule inhibitors of PD-1/PD-L1 immune checkpoint alleviate the PD-L1-induced exhaustion of T-cells. Oncotarget 2017, 8, 72167–72181. [Google Scholar] [CrossRef] [Green Version]
- Shaabani, S.; Huizinga, H.P.S.; Butera, R.; Kouchi, A.; Guzik, K.; Magiera-Mularz, K.; Holak, T.A.; Dömling, A. A patent review on PD-1/PD-L1 antagonists: Small molecules, peptides, and macrocycles (2015–2018). Expert. Opin. Ther. Pat. 2018, 28, 665–678. [Google Scholar] [CrossRef] [Green Version]
- Shi, D.; An, X.; Bai, Q.; Bing, Z.; Zhou, S.; Liu, H.; Yao, X. Computational insight into the small molecule intervening PD-L1 dimerization and the potential structure-activity relationship. Front. Chem. 2019, 7, 764. [Google Scholar] [CrossRef] [Green Version]
- Ganesan, A.; Ahmed, M.; Okoye, I.; Arutyunova, E.; Babu, D.; Turnbull, W.L.; Kundu, J.K.; Shields, J.; Agopsowicz, K.C.; Xu, L.; et al. Comprehensive in vitro characterization of PD-L1 small molecule inhibitors. Sci. Rep. 2019, 9, 12392. [Google Scholar] [CrossRef]
- Guo, Y.; Jin, Y.; Wang, B.; Liu, B. Molecular mechanism of small-molecule inhibitors in blocking the PD-1/PD-L1 pathway through PD-L1 dimerization. Int. J. Mol. Sci. 2021, 22, 4766. [Google Scholar] [CrossRef]
- Dai, X.; Wang, K.; Chen, H.; Huang, X.; Feng, Z. Design, synthesis, and biological evaluation of 1-methyl-1H-pyrazolo[4,3-b]pyridine derivatives as novel small-molecule inhibitors targeting the PD-1/PD-L1 interaction. Bioorg. Chem. 2021, 114, 105034. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, K.; Chen, H.; Feng, Z. Design, synthesis, evaluation, and SAR of 4-phenylindoline derivatives, a novel class of small-molecule inhibitors of the programmed cell death-1/ programmed cell death-ligand 1 (PD-1/PD-L1) interaction. Eur. J. Med. Chem. 2021, 211, 113001. [Google Scholar] [CrossRef]
- Russomanno, P.; Assoni, G.; Amato, J.; D’Amore, V.M.; Scaglia, R.; Brancaccio, D.; Pedrini, M.; Polcaro, G.; La Pietra, V.; Orlando, P.; et al. Interfering with the tumor-immune interface: Making way for triazine-based small molecules as novel PD-L1 inhibitors. J. Med. Chem. 2021, 64, 16020–16045. [Google Scholar] [CrossRef]
- Zhang, H.; Xia, Y.; Yu, C.; Du, H.; Liu, J.; Li, H.; Huang, S.; Zhu, Q.; Xu, Y.; Zou, Y. Discovery of novel small-molecule inhibitors of PD-1/PD-L1 interaction via structural simplification strategy. Molecules 2021, 26, 3347. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Yuan, D.; Ma, J. Advances of biphenyl small-molecule inhibitors targeting PD-1/PD-L1 interaction in cancer immunotherapy. Future Med. Chem. 2022, 14, 97–113. [Google Scholar] [CrossRef]
- Kopalli, S.R.; Kang, T.B.; Lee, K.H.; Koppula, S. Novel small molecule inhibitors of programmed cell death (PD)-1, and its ligand, PD-L1 in cancer immunotherapy: A review update of patent literature. Recent Pat. Anticancer Drug Discov. 2019, 14, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Thuru, X.; Quesnel, M.; Magnez, R.; Millet, R.; Leleu, N.; Klupsch, F. Novel pyrazolone derivatives as PD-1/PD-L1 interaction inhibitors. EP3766544, PCT/EP2020/070478 (WO/2021/009384).
- Le Biannic, R.; Magnez, R.; Klupsch, F.; Leleu-Chavain, N.; Thiroux, B.; Tardy, M.; El Bouazzati, H.; Dezitter, X.; Renault, N.; Vergoten, G.; et al. Pyrazolones as inhibitors of immune checkpoint blocking the PD-1/PD-L1 interaction. Eur. J. Med. Chem. 2022, 236, 114343. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, H. Edaravone for the treatment of amyotrophic lateral sclerosis. Expert Rev. Neurother. 2019, 19, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Li, M.; Lin, L.; Chen, S.; Chen, Y.; Hong, L. Clinical effects and safety of edaravone in treatment of acute ischaemic stroke: A meta-analysis of randomized controlled trials. J. Clin. Pharm. Ther. 2021, 46, 907–917. [Google Scholar] [CrossRef]
- Bailly, C. Potential use of edaravone to reduce specific side effects of chemo-, radio- and immuno-therapy of cancers. Int. Immunopharmacol. 2019, 77, 105967. [Google Scholar] [CrossRef]
- Cha, S.J.; Kim, K. Effects of the edaravone, a drug approved for the treatment of amyotrophic lateral sclerosis, on mitochondrial function and neuroprotection. Antioxidants 2022, 11, 195. [Google Scholar] [CrossRef]
- Ren, Y.; Wei, B.; Song, X.; An, N.; Zhou, Y.; Jin, X.; Zhang, Y. Edaravone’s free radical scavenging mechanisms of neuroprotection against cerebral ischemia: Review of the literature. Int. J. Neurosci. 2015, 125, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Ahmadinejad, F.; Geir Møller, S.; Hashemzadeh-Chaleshtori, M.; Bidkhori, G.; Jami, M.S. Molecular mechanisms behind free radical scavengers function against oxidative stress. Antioxidants 2017, 6, 51. [Google Scholar] [CrossRef]
- Arai, T.; Nonogawa, M.; Makino, K.; Endo, N.; Mori, H.; Miyoshi, T.; Yamashita, K.; Sasada, M.; Kakuyama, M.; Fukuda, K. The radical scavenger edaravone (3-methyl-1-phenyl-2-pyrazolin-5-one) reacts with a pterin derivative and produces a cytotoxic substance that induces intracellular reactive oxygen species generation and cell death. J. Pharmacol. Exp. Ther. 2008, 324, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Nonogawa, M.; Arai, T.; Endo, N.; Pack, S.P.; Kodaki, T.; Makino, K. Reactive oxygen species generation through NADH oxidation by pterin derivatives. Nucleic Acids Symp. Ser. 2008, 52, 567–568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goossens, J.F.; Thuru, X.; Bailly, C. Properties and reactivity of the folic acid and folate photoproduct 6-formylpterin. Free Radic. Biol. Med. 2021, 171, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bailly, C.; Hecquet, P.E.; Kouach, M.; Thuru, X.; Goossens, J.F. Chemical reactivity and uses of 1-phenyl-3-methyl-5-pyrazolone (PMP), also known as edaravone. Bioorg. Med. Chem. 2020, 28, 115463. [Google Scholar] [CrossRef]
- Regnault, R.; Kouach, M.; Goossens, L.; Thuru, X.; Bailly, C.; Goossens, J.F. Mono- and bis-edaravone adducts formed in the presence of vanillin in an aqueous solution. Sep. Sci. Plus 2022, 1–12. [Google Scholar] [CrossRef]
- Stanojević, L.; Stanković, M.; Nikolić, V.; Nikolić, L.; Ristić, D.; Čanadanovic-Brunet, J.; Tumbas, V. Antioxidant activity and total phenolic and flavonoid contents of Hieracium pilosella L. Extr. Sens. 2009, 9, 5702–5714. [Google Scholar] [CrossRef] [Green Version]
- Pasanphan, W.; Buettner, G.R.; Chirachanchai, S. Chitosan gallate as a novel potential polysaccharide antioxidant: An EPR study. Carbohydr. Res. 2010, 345, 132–140. [Google Scholar] [CrossRef] [Green Version]
- Kamogawa, E.; Sueishi, Y. A multiple free-radical scavenging (MULTIS) study on the antioxidant capacity of a neuroprotective drug, edaravone as compared with uric acid, glutathione, and trolox. Bioorg. Med. Chem. Lett. 2014, 24, 1376–1379. [Google Scholar] [CrossRef]
- Abe, S.; Kirima, K.; Tsuchiya, K.; Okamoto, M.; Hasegawa, T.; Houchi, H.; Yoshizumi, M.; Tamaki, T. The reaction rate of edaravone (3-methyl-1-phenyl-2-pyrazolin-5-one (MCI-186)) with hydroxyl radical. Chem. Pharm. Bull. 2004, 52, 186–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morehouse, K.M.; Mason, R.P. The transition metal-mediated formation of the hydroxyl free radical during the reduction of molecular oxygen by ferredoxin-ferredoxin:NADP+ oxidoreductase. J. Biol. Chem. 1988, 263, 1204–1211. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X.; Zou, G.; Peng, S.; Liu, C.; Zhou, X. Detection and Application of 5-Formylcytosine and 5-Formyluracil in DNA. Acc. Chem. Res. 2019, 52, 1016–1024. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Han, S.; Zhang, X.; Wang, Y.; Zou, G.; Liu, C.; Xu, M.; Zhou, X. Sequencing 5-formyluracil in genomic DNA at single-base resolution. Anal. Chem. 2021, 93, 15445–15451. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Shen, S.; Xing, S.; Yu, H.; Huan, T. ISFrag: De novo recognition of in-source fragments for liquid chromatography-mass spectrometry data. Anal. Chem. 2021, 93, 10243–10250. [Google Scholar] [CrossRef] [PubMed]
- Kono, H.; Woods, C.G.; Maki, A.; Connor, H.D.; Mason, R.P.; Rusyn, I.; Fujii, H. Electron spin resonance and spin trapping technique provide direct evidence that edaravone prevents acute ischemia-reperfusion injury of the liver by limiting free radical-mediated tissue damage. Free Radic. Res. 2006, 40, 579–588. [Google Scholar] [CrossRef]
- Hata, K.; Lin, M.; Katsumura, Y.; Muroya, Y.; Fu, H.; Yamashita, S.; Nakagawa, H. Pulse radiolysis study on free radical scavenger edaravone (3-methyl-1-phenyl-2-pyrazolin-5-one). 2: A comparative study on edaravone derivatives. J. Radiat. Res. 2011, 52, 15–23. [Google Scholar] [CrossRef] [Green Version]
- Bailly, C. Regulation of PD-L1 expression on cancer cells with ROS-modulating drugs. Life Sci. 2020, 246, 117403. [Google Scholar] [CrossRef]
- Kongtawelert, P.; Wudtiwai, B.; Shwe, T.H.; Pothacharoen, P.; Phitak, T. Inhibition of programmed death ligand 1 (PD-L1) expression in breast cancer cells by sesamin. Int. Immunopharmacol. 2020, 86, 106759. [Google Scholar] [CrossRef]
- Sp, N.; Kang, D.Y.; Lee, J.M.; Jang, K.J. Mechanistic Insights of Anti-Immune Evasion by Nobiletin through Regulating miR-197/STAT3/PD-L1 Signaling in Non-Small Cell Lung Cancer (NSCLC) Cells. Int. J. Mol. Sci. 2021, 22, 9843. [Google Scholar] [CrossRef]
- Mendonca, P.; Hill, L.; Soliman, K.F.A. Effects of Cardamonin on PD-1/PD-L1 Checkpoint in Triple-Negative Breast Cancer. FASEB J. 2022, 36 (Suppl. 1). [Google Scholar] [CrossRef]
- Evans, J.; Mendonca, P.; Soliman, K.F.A. Hesperetin modulation of oxidative stress and inflammatory mediators in LPS-Activated BV-2 Microglial Cells. FASEB J. 2022, 36 (Suppl. 1). [Google Scholar] [CrossRef]
- An, E.K.; Hwang, J.; Kim, S.J.; Park, H.B.; Zhang, W.; Ryu, J.H.; You, S.; Jin, J.O. Comparison of the immune activation capacities of fucoidan and laminarin extracted from Laminaria japonica. Int. J. Biol. Macromol. 2022, 208, 230–242. [Google Scholar] [CrossRef] [PubMed]
- Peng, D.; He, A.; He, S.; Ge, G.; Wang, S.; Ci, W.; Li, X.; Xia, D.; Zhou, L. Ascorbic acid induced TET2 enzyme activation enhances cancer immunotherapy efficacy in renal cell carcinoma. Int. J. Biol. Sci. 2022, 18, 995–1007. [Google Scholar] [CrossRef] [PubMed]
- Zuo, H.X.; Jin, Y.; Wang, Z.; Li, M.Y.; Zhang, Z.H.; Wang, J.Y.; Xing, Y.; Ri, M.H.; Jin, C.H.; Xu, G.H.; et al. Curcumol inhibits the expression of programmed cell death-ligand 1 through crosstalk between hypoxia-inducible factor-1α and STAT3 (T705) signaling pathways in hepatic cancer. J. Ethnopharmacol. 2020, 257, 112835. [Google Scholar] [CrossRef]
- Fu, J.; Wu, Z.; Liu, J.; Wu, T. Vitamin C: A stem cell promoter in cancer metastasis and immunotherapy. Biomed. Pharmacother. 2020, 131, 110588. [Google Scholar] [CrossRef]
- Bedhiafi, T.; Inchakalody, V.P.; Fernandes, Q.; Mestiri, S.; Billa, N.; Uddin, S.; Merhi, M.; Dermime, S. The potential role of vitamin C in empowering cancer immunotherapy. Biomed. Pharmacother. 2022, 146, 112553. [Google Scholar] [CrossRef]
- Bakalova, R.; Semkova, S.; Ivanova, D.; Zhelev, Z.; Miller, T.; Takeshima, T.; Shibata, S.; Lazarova, D.; Aoki, I.; Higashi, T. Selective Targeting of Cancerous Mitochondria and Suppression of Tumor Growth Using Redox-Active Treatment Adjuvant. Oxid. Med. Cell Longev. 2020, 2020, 6212935. [Google Scholar] [CrossRef]
- Glorieux, C.; Xia, X.; He, Y.Q.; Hu, Y.; Cremer, K.; Robert, A.; Liu, J.; Wang, F.; Ling, J.; Chiao, P.J.; et al. Regulation of PD-L1 expression in K-ras-driven cancers through ROS-mediated FGFR1 signaling. Redox Biol. 2021, 38, 101780. [Google Scholar] [CrossRef]
- Luo, D.; Bai, H.; Zhou, X.; Wu, L.; Zhang, C.; Wu, Z.; Li, Z.; Bai, L. Synthesis of herbicide candidate through optimization of quinclorac with 3-methyl-1H-pyrazol-5-yl. Front. Chem. 2021, 9, 647472. [Google Scholar] [CrossRef]
- Magnez, R.; Thiroux, B.; Taront, S.; Segaoula, Z.; Quesnel, B.; Thuru, X. PD-1/PD-L1 binding studies using microscale thermophoresis. Sci. Rep. 2017, 7, 17623. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Compounds | PD-L1 Binding | Bioactivity | Proliferation |
|---|---|---|---|
| (KD, nM) a | (IC50, nM) b | (IC50, nM) c | |
| 1 | 77 ± 7 | 92 ± 9 | 102 ± 8 |
| 2 | 34 ± 3 | 23 ± 3 | 53 ± 11 |
| 3 | 45 ± 7 | 44 ± 9 | 79 ± 7 |
| 4 | 7 ± 3 | 3 ± 2 | 124 ± 7 |
| 5 | 12 ± 2 | 23 ± 6 | 57 ± 9 |
| BMS-202 | ND | 124 ± 12 | 53 ± 17 |
| Nivolumab | ND | ND | 58 ± 3 |
| Cpd | DMPO | DPPH | |
|---|---|---|---|
| kr (1011 M−1 s−1) | kr/ka | EC50 (μM) | |
| EDA | 2.59 | 60 | 35.81 |
| 1 | 1.27 | 30 | 70.58 |
| 2 | 1.93 | 45 | 36.46 |
| 3 | 1.88 | 44 | 34.02 |
| 4 | 1.04 | 24 | 97.58 |
| 5 | 1.25 | 29 | 59.74 |
| Analyte [M-H]− | Formula | Ions m/z(Theorical) | Ions m/z(Observed) | Error Δppm | RDB Negative Mode |
|---|---|---|---|---|---|
| 5fU | C5H3N2O3 | 139.022 | 139.013 | −3.946 | 5.5 |
| EDA | C10H9N2O | 173.080 | 173.070 | −2.713 | 7.5 |
| U-(EDA)1 | C15H11N4O3 | 295.090 | 295.082 | −1.141 | 12.5 |
| U-(EDA)2 | C25H21N6O4 | 469.170 | 469.161 | −1.960 | 18.5 |
| Cpd 1 | C9H8N3O | 174.075 | 174.066 | −2.576 | 7.5 |
| U-(1)1 | C14H10N5O3 | 296.086 | 296.078 | −0.627 | 12.5 |
| U-(1)2 | C23H19N8O4 | 471.160 | 471.152 | −1.799 | 18.5 |
| Cpd 2 | C10H7N2OCl2 | 241.001 | 240.993 | −0.269 | 7.5 |
| U-(2)1 | C15H9N4O3Cl2 | 363.013 | 363.004 | −0.639 | 12.5 |
| U-(2)2 | C25H17N6O4Cl4 | 605.014 | 605.005 | −2.149 | 18.5 |
| Cpd 3 | C16H11N2O2Cl2 | 333.028 | 333.019 | −0.299 | 11.5 |
| U-(3)1 | C15H9N4O3Cl2 | 455.039 | 455.030 | −1.421 | 16.5 |
| U-(3)2 | C25H17N6O4Cl4 | 789.067 | 789.056 | −2.648 | 26.5 |
| Cpd 4 | C15H8N2OCl3 | 336.977 | 336.969 | −2.352 | 11.5 |
| U-(4)1 | C20H10N4O3Cl3 | 458.990 | 458.980 | −3.507 | 16.5 |
| U-(4)2 | C35H19N6O4Cl6 | 796.68 | 796.956 | −4.612 | 26.5 |
| Cpd 5 | C19H20N2OCl | 327.134 | 327.125 | −1.704 | 10.5 |
| U-(5)1 | C24H22N4O3Cl | 449.146 | No reaction | ||
| U-(5)2 | C43H43N6O4Cl2 | 777.280 | No reaction |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leleu-Chavain, N.; Regnault, R.; Ahouari, H.; Le Biannic, R.; Kouach, M.; Klupsch, F.; Magnez, R.; Vezin, H.; Thuru, X.; Bailly, C.; et al. Antioxidant Properties and Aldehyde Reactivity of PD-L1 Targeted Aryl-Pyrazolone Anticancer Agents. Molecules 2022, 27, 3316. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27103316
Leleu-Chavain N, Regnault R, Ahouari H, Le Biannic R, Kouach M, Klupsch F, Magnez R, Vezin H, Thuru X, Bailly C, et al. Antioxidant Properties and Aldehyde Reactivity of PD-L1 Targeted Aryl-Pyrazolone Anticancer Agents. Molecules. 2022; 27(10):3316. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27103316
Chicago/Turabian StyleLeleu-Chavain, Natascha, Romain Regnault, Hania Ahouari, Raphaël Le Biannic, Mostafa Kouach, Frédérique Klupsch, Romain Magnez, Hervé Vezin, Xavier Thuru, Christian Bailly, and et al. 2022. "Antioxidant Properties and Aldehyde Reactivity of PD-L1 Targeted Aryl-Pyrazolone Anticancer Agents" Molecules 27, no. 10: 3316. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27103316