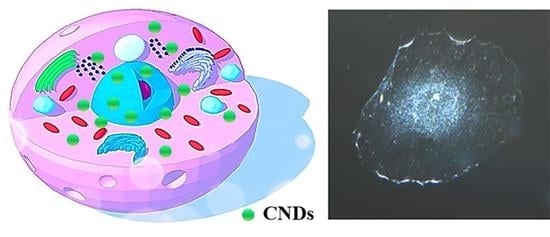
Dark-Field Microscopic Study of Cellular Uptake of Carbon Nanodots: Nuclear Penetrability
Abstract
:1. Introduction
2. Results
2.1. Synthesis and Characterization of CNDs
2.2. Dark-Field Microscopy and Hyperspectral Imaging of Cells with CND Incubation at Different Time
2.3. Light Intensity Measurements under CNDs Incubation with Different Concentrations
3. Discussion
4. Materials and Methods
4.1. Synthesis of CNDs
4.2. Characterization of CNDs
4.3. Cell Culture
4.4. Sample Fixation
4.5. Dark-Field Microscopy with Hyperspectral Imaging
4.6. 3-(4,5-Dimethylthiazol-2yl)-2,5-diphenyltetrazolium Bromide (MTT)-Based Assay
4.7. Confocal Imaging
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Zhu, S.; Meng, Q.; Wang, L.; Zhang, J.; Song, Y.; Jin, H.; Zhang, K.; Sun, H.; Wang, H.; Yang, B. Highly Photoluminescent Carbon Dots for Multicolor Patterning, Sensors, and Bioimaging. Angew. Chem. Int. Ed. 2013, 52, 3953–3957. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.Y.; Shen, W.; Gao, Z. Carbon quantum dots and their applications. Chem. Soc. Rev. 2015, 44, 362–381. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Kang, Z.; Liu, Y.; Lee, S.-T. Carbon nanodots: Synthesis, properties and applications. J. Mater. Chem. 2012, 22, 24230–24253. [Google Scholar] [CrossRef]
- Roy, P.; Chen, P.-C.; Periasamy, A.P.; Chen, Y.-N.; Chang, H.-T. Photoluminescent carbon nanodots: Synthesis, physicochemical properties and analytical applications. Mater. Today 2015, 18, 447–458. [Google Scholar] [CrossRef]
- Kumar, V.B.; Perelshtein, I.; Lipovsky, A.; Porat, Z.E.; Gedanken, A. The sonochemical synthesis of Ga@C-dots particles. RSC Adv. 2015, 5, 25533–25540. [Google Scholar] [CrossRef]
- Kumar, V.B.; Porat, Z.E.; Gedanken, A. Facile one-step sonochemical synthesis of ultrafine and stable fluorescent C-dots. Ultrason. Sonochem. 2016, 28, 367–375. [Google Scholar] [CrossRef]
- Bagra, B.; Zhang, W.; Zeng, Z.; Mabe, T.; Wei, J. Plasmon-Enhanced Fluorescence of Carbon Nanodots in Gold Nanoslit Cavities. Langmuir 2019, 35, 8903–8909. [Google Scholar] [CrossRef]
- Arvapalli, D.M.; Sheardy, A.T.; Alapati, K.C.; Wei, J. High Quantum Yield Fluorescent Carbon Nanodots for detection of Fe (III) Ions and Electrochemical Study of Quenching Mechanism. Talanta 2020, 209, 120538. [Google Scholar] [CrossRef]
- Liu, J.; Li, D.; Zhang, K.; Yang, M.; Sun, H.; Yang, B. One-Step Hydrothermal Synthesis of Nitrogen-Doped Conjugated Carbonized Polymer Dots with 31% Efficient Red Emission for In Vivo Imaging. Small 2018, 14, 1703919. [Google Scholar] [CrossRef]
- Liu, J.; Lu, S.; Tang, Q.; Zhang, K.; Yu, W.; Sun, H.; Yang, B. One-step hydrothermal synthesis of photoluminescent carbon nanodots with selective antibacterial activity against Porphyromonas gingivalis. Nanoscale 2017, 9, 7135–7142. [Google Scholar] [CrossRef]
- Zheng, X.T.; Ananthanarayanan, A.; Luo, K.Q.; Chen, P. Glowing Graphene Quantum Dots and Carbon Dots: Properties, Syntheses, and Biological Applications. Small 2014, 11, 1620–1636. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hu, A. Carbon quantum dots: Synthesis, properties and applications. J. Mater. Chem. C 2014, 2, 6921–6939. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Z.; Zhang, W.; Arvapalli, D.M.; Bloom, B.; Sheardy, A.; Mabe, T.; Liu, Y.; Ji, Z.; Chevva, H.; Waldeck, D.H.; et al. A fluorescence-electrochemical study of carbon nanodots (CNDs) in bio- and photoelectronic applications and energy gap investigation. Phys. Chem. Chem. Phys. 2017, 19, 20101–20109. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.-Y.; Gedda, G.; Girma, W.M.; Yen, C.-L.; Ling, Y.-C.; Chang, J.-Y. Magnetofluorescent Carbon Dots Derived from Crab Shell for Targeted Dual-Modality Bioimaging and Drug Delivery. ACS Appl. Mater. Interfaces 2017, 9, 13887–13899. [Google Scholar] [CrossRef]
- Cheng, W.; Xu, J.; Guo, Z.; Yang, D.; Chen, X.; Yan, W.; Miao, P. Hydrothermal synthesis of N,S co-doped carbon nanodots for highly selective detection of living cancer cells. J. Mater. Chem. B 2018, 6, 5775–5780. [Google Scholar] [CrossRef]
- Miao, P.; Han, K.; Tang, Y.; Wang, B.; Lin, T.; Cheng, W. Recent advances in carbon nanodots: Synthesis, properties and biomedical applications. Nanoscale 2015, 7, 1586–1595. [Google Scholar] [CrossRef]
- Chen, T.-H.; Tseng, W.-L. Self-Assembly of Monodisperse Carbon Dots into High-Brightness Nanoaggregates for Cellular Uptake Imaging and Iron(III) Sensing. Anal. Chem. 2017, 89, 11348–11356. [Google Scholar] [CrossRef]
- Ji, Z.; Arvapalli, D.M.; Zhang, W.; Yin, Z.; Wei, J. Nitrogen and sulfur co-doped carbon nanodots in living EA.hy926 and A549 cells: Oxidative stress effect and mitochondria targeting. J. Mater. Sci. 2020, 55, 6093–6104. [Google Scholar] [CrossRef]
- Ji, Z.; Sheardy, A.; Zeng, Z.; Zhang, W.; Chevva, H.; Allado, K.; Yin, Z.; Wei, J. Tuning the Functional Groups on Carbon Nanodots and Antioxidant Studies. Molecules 2019, 24, 152. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Zeng, Z.; Wei, J. Electrochemical Study of DPPH Radical Scavenging for Evaluating the Antioxidant Capacity of Carbon Nanodots. J. Phys. Chem. C 2017, 121, 18635–18642. [Google Scholar] [CrossRef]
- Arvapalli, D.M.; Sheardy, A.T.; Allado, K.; Chevva, H.; Yin, Z.; Wei, J. Design of Curcumin Loaded Carbon Nanodots Delivery System: Enhanced Bioavailability, Release Kinetics, and Anticancer Activity. ACS Appl. Bio Mater. 2020, 3, 8776–8785. [Google Scholar] [CrossRef] [PubMed]
- Arvapalli, D.M.; Sheardy, A.T.; Bang, J.J.; Wei, J. Antiproliferative and ROS Regulation Activity of Photoluminescent Curcumin-Derived Nanodots. ACS Appl. Bio Mater. 2021, 4, 8477–8486. [Google Scholar] [CrossRef] [PubMed]
- Dunphy, A.; Patel, K.; Belperain, S.; Pennington, A.; Chiu, N.H.L.; Yin, Z.; Zhu, X.; Priebe, B.; Tian, S.; Wei, J.; et al. Modulation of Macrophage Polarization by Carbon Nanodots and Elucidation of Carbon Nanodot Uptake Routes in Macrophages. Nanomaterials 2021, 11, 1116. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-F.; Wu, H.-C.; Kuan, C.-H.; Lin, C.-J.; Wang, L.-W.; Chang, C.-W.; Wang, T.-W. Multi-functionalized carbon dots as theranostic nanoagent for gene delivery in lung cancer therapy. Sci. Rep. 2016, 6, 21170. [Google Scholar] [CrossRef]
- Khan, S.; Verma, N.C.; Chethana; Nandi, C.K. Carbon Dots for Single-Molecule Imaging of the Nucleolus. ACS Appl. Nano Mater. 2018, 1, 483–487. [Google Scholar] [CrossRef]
- Dekaliuk, M.; Pyrshev, K.; Demchenko, A. Visualization and detection of live and apoptotic cells with fluorescent carbon nanoparticles. J. Nanobiotechnol. 2015, 13, 86. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Neshatian, M.; van Prooijen, M. Cancer Nanotechnology: Enhanced Therapeutic Response Using Peptide-Modified Gold Nanoparticles. J. Nanosci. Nanotechnol. 2014, 14, 4813–4819. [Google Scholar] [CrossRef]
- Shukla, R.; Bansal, V.; Chaudhary, M.; Basu, A.; Bhonde, R.R.; Sastry, M. Biocompatibility of Gold Nanoparticles and Their Endocytotic Fate Inside the Cellular Compartment: A Microscopic Overview. Langmuir 2005, 21, 10644–10654. [Google Scholar] [CrossRef]
- Abadeer, N.S.; Murphy, C.J. Recent Progress in Cancer Thermal Therapy Using Gold Nanoparticles. J. Phys. Chem. C 2016, 120, 4691–4716. [Google Scholar] [CrossRef]
- Ma, X.; Hartmann, R.; Jimenez de Aberasturi, D.; Yang, F.; Soenen, S.J.H.; Manshian, B.B.; Franz, J.; Valdeperez, D.; Pelaz, B.; Feliu, N.; et al. Colloidal Gold Nanoparticles Induce Changes in Cellular and Subcellular Morphology. ACS Nano 2017, 11, 7807–7820. [Google Scholar] [CrossRef]
- Dimitriou, N.M.; Tsekenis, G.; Balanikas, E.C.; Pavlopoulou, A.; Mitsiogianni, M.; Mantso, T.; Pashos, G.; Boudouvis, A.G.; Lykakis, I.N.; Tsigaridas, G.; et al. Gold nanoparticles, radiations and the immune system: Current insights into the physical mechanisms and the biological interactions of this new alliance towards cancer therapy. Pharmacol. Ther. 2017, 178, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Li, M.; Chen, B.; Shen, Z.; Guo, P.; Wientjes, M.G.; Au, J.L.S. Predictive Models of Diffusive Nanoparticle Transport in 3-Dimensional Tumor Cell Spheroids. AAPS J. 2013, 15, 816–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.-D.; Wu, D.; Shen, X.; Chen, J.; Sun, Y.-M.; Liu, P.-X.; Liang, X.-J. Size-dependent radiosensitization of PEG-coated gold nanoparticles for cancer radiation therapy. Biomaterials 2012, 33, 6408–6419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, L.; Shen, J.; Li, Z.; Zhang, D.; Zhang, Q.; Duan, C.; Liu, G.; Zheng, D.; Liu, Y.; Tian, X. Successfully tailoring the pore size of mesoporous silica nanoparticles: Exploitation of delivery systems for poorly water-soluble drugs. Int. J. Pharm. 2012, 439, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Yameen, B.; Choi, W.I.; Vilos, C.; Swami, A.; Shi, J.; Farokhzad, O.C. Insight into nanoparticle cellular uptake and intracellular targeting. J. Control. Release 2014, 190, 485–499. [Google Scholar] [CrossRef] [Green Version]
- Jin, H.; Heller, D.A.; Sharma, R.; Strano, M.S. Size-Dependent Cellular Uptake and Expulsion of Single-Walled Carbon Nanotubes: Single Particle Tracking and a Generic Uptake Model for Nanoparticles. ACS Nano 2009, 3, 149–158. [Google Scholar] [CrossRef]
- Combs, C.A. Fluorescence Microscopy: A Concise Guide to Current Imaging Methods. Curr. Protoc. Neurosci. 2010, 50, 2.1.1–2.1.14. [Google Scholar] [CrossRef]
- Papaioannou, N.; Marinovic, A.; Yoshizawa, N.; Goode, A.E.; Fay, M.; Khlobystov, A.; Titirici, M.-M.; Sapelkin, A. Structure and solvents effects on the optical properties of sugar-derived carbon nanodots. Sci. Rep. 2018, 8, 6559. [Google Scholar] [CrossRef]
- Lu, S.; Sui, L.; Liu, J.; Zhu, S.; Chen, A.; Jin, M.; Yang, B. Near-Infrared Photoluminescent Polymer–Carbon Nanodots with Two-Photon Fluorescence. Adv. Mater. 2017, 29, 1603443. [Google Scholar] [CrossRef]
- Ong, W.-J.; Putri, L.K.; Tan, Y.-C.; Tan, L.-L.; Li, N.; Ng, Y.H.; Wen, X.; Chai, S.-P. Unravelling charge carrier dynamics in protonated g-C3N4 interfaced with carbon nanodots as co-catalysts toward enhanced photocatalytic CO2 reduction: A combined experimental and first-principles DFT study. Nano Res. 2017, 10, 1673–1696. [Google Scholar] [CrossRef]
- Chizhik, A.M.; Stein, S.; Dekaliuk, M.O.; Battle, C.; Li, W.; Huss, A.; Platen, M.; Schaap, I.A.T.; Gregor, I.; Demchenko, A.P.; et al. Super-Resolution Optical Fluctuation Bio-Imaging with Dual-Color Carbon Nanodots. Nano Lett. 2016, 16, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.K.; Shin, E.; Kim, B.-S. Cell Nucleus-Targeting Zwitterionic Carbon Dots. Sci. Rep. 2015, 5, 18807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hua, X.-W.; Bao, Y.-W.; Chen, Z.; Wu, F.-G. Carbon quantum dots with intrinsic mitochondrial targeting ability for mitochondria-based theranostics. Nanoscale 2017, 9, 10948–10960. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Jiang, Y.-W.; Yang, J.; Wu, F.-G. Mitochondria-targetable carbon quantum dots for differentiating cancerous cells from normal cells. Nanoscale 2017, 9, 18368–18378. [Google Scholar] [CrossRef]
- Zhang, W.; Chavez, J.; Zeng, Z.; Bloom, B.; Sheardy, A.; Ji, Z.; Yin, Z.; Waldeck, D.H.; Jia, Z.; Wei, J. Antioxidant Capacity of Nitrogen and Sulfur Codoped Carbon Nanodots. ACS Appl. Nano Mater. 2018, 1, 2699–2708. [Google Scholar] [CrossRef]
- Kasouni, A.; Chatzimitakos, T.; Stalikas, C. Bioimaging Applications of Carbon Nanodots: A Review. C-J. Carbon Res. 2019, 5, 19. [Google Scholar] [CrossRef] [Green Version]
- Hua, X.-W.; Bao, Y.-W.; Wu, F.-G. Fluorescent Carbon Quantum Dots with Intrinsic Nucleolus-Targeting Capability for Nucleolus Imaging and Enhanced Cytosolic and Nuclear Drug Delivery. ACS Appl. Mater. Interfaces 2018, 10, 10664–10677. [Google Scholar] [CrossRef]
- Elsaesser, A.; Taylor, A.; de Yanés, G.S.; McKerr, G.; Kim, E.-M.; O’Hare, E.; Howard, C.V. Quantification of nanoparticle uptake by cells using microscopical and analytical techniques. Nanomedicine 2010, 5, 1447–1457. [Google Scholar] [CrossRef]
- Wolfbeis, O.S. An overview of nanoparticles commonly used in fluorescent bioimaging. Chem. Soc. Rev. 2015, 44, 4743–4768. [Google Scholar] [CrossRef] [Green Version]
- Ivask, A.; Mitchell, A.J.; Malysheva, A.; Voelcker, N.H.; Lombi, E. Methodologies and approaches for the analysis of cell–nanoparticle interactions. WIREs Nanomed. Nanobiotechnol. 2017, 10, e1486. [Google Scholar] [CrossRef]
- Yguerabide, J.; Yguerabide, E.E. Light-Scattering Submicroscopic Particles as Highly Fluorescent Analogs and Their Use as Tracer Labels in Clinical and Biological Applications: I. Theory. Anal. Biochem. 1998, 262, 137–156. [Google Scholar] [CrossRef] [PubMed]
- Dai, Q.; Liu, X.; Coutts, J.; Austin, L.; Huo, Q. A One-Step Highly Sensitive Method for DNA Detection Using Dynamic Light Scattering. J. Am. Chem. Soc. 2008, 130, 8138–8139. [Google Scholar] [CrossRef] [PubMed]
- Zamora-Perez, P.; Tsoutsi, D.; Xu, R.; Rivera_Gil, P. Hyperspectral-Enhanced Dark Field Microscopy for Single and Collective Nanoparticle Characterization in Biological Environments. Materials 2018, 11, 243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Essner, J.B.; Kist, J.A.; Polo-Parada, L.; Baker, G.A. Artifacts and Errors Associated with the Ubiquitous Presence of Fluorescent Impurities in Carbon Nanodots. Chem. Mater. 2018, 30, 1878–1887. [Google Scholar] [CrossRef]
- Li, W.; Zhang, Z.; Kong, B.; Feng, S.; Wang, J.; Wang, L.; Yang, J.; Zhang, F.; Wu, P.; Zhao, D. Simple and Green Synthesis of Nitrogen-Doped Photoluminescent Carbonaceous Nanospheres for Bioimaging. Angew. Chem. Int. Ed. 2013, 52, 8151–8155. [Google Scholar] [CrossRef]
- Oh, E.S.; Heo, C.; Kim, J.S.; Suh, M.; Lee, Y.H.; Kim, J.-M. Hyperspectral fluorescence imaging for cellular iron mapping in the in vitro model of Parkinson’s disease. J. Biomed. Opt. 2013, 19, 051207. [Google Scholar] [CrossRef] [Green Version]
- Zhu, S.; Song, Y.; Zhao, X.; Shao, J.; Zhang, J.; Yang, B. The photoluminescence mechanism in carbon dots (graphene quantum dots, carbon nanodots, and polymer dots): Current state and future perspective. Nano Res. 2015, 8, 355–381. [Google Scholar] [CrossRef]
- Yang, C.; Uertz, J.; Yohan, D.; Chithrani, B.D. Peptide modified gold nanoparticles for improved cellular uptake, nuclear transport, and intracellular retention. Nanoscale 2014, 6, 12026–12033. [Google Scholar] [CrossRef]
- Mao, Q.-X.; E, S.; Xia, J.-M.; Song, R.-S.; Shu, Y.; Chen, X.-W.; Wang, J.-H. Hydrophobic Carbon Nanodots with Rapid Cell Penetrability and Tunable Photoluminescence Behavior for in Vitro and in Vivo Imaging. Langmuir 2016, 32, 12221–12229. [Google Scholar] [CrossRef]
- Puri, V.; Watanabe, R.; Singh, R.D.; Dominguez, M.; Brown, J.C.; Wheatley, C.L.; Marks, D.L.; Pagano, R.E. Clathrin-dependent and-independent internalization of plasma membrane sphingolipids initiates two Golgi targeting pathways. J. Cell Biol. 2001, 154, 535. [Google Scholar] [CrossRef] [Green Version]
- Zhou, N.; Zhu, S.; Maharjan, S.; Hao, Z.; Song, Y.; Zhao, X.; Jiang, Y.; Yang, B.; Lu, L. Elucidating the endocytosis, intracellular trafficking, and exocytosis of carbon dots in neural cells. RSC Adv. 2014, 4, 62086–62095. [Google Scholar] [CrossRef]
- Fernandez-Martinez, J.; Rout, M. Nuclear pore complex biogenesis. Curr. Opin. Cell Biol. 2009, 21, 603–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suntharalingam, M.; Wente, S.R. Peering through the Pore: Nuclear Pore Complex Structure, Assembly, and Function. Dev. Cell 2003, 4, 775–789. [Google Scholar] [CrossRef]
- Belli, V.; Guarnieri, D.; Biondi, M.; della Sala, F.; Netti, P.A. Dynamics of nanoparticle diffusion and uptake in three-dimensional cell cultures. Colloids Surf. B 2017, 149, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Goodman, T.T.; Chen, J.; Matveev, K.; Pun, S.H. Spatio-Temporal Modeling of Nanoparticle Delivery to Multicellular Tumor Spheroids. Biotechnol. Bioeng. 2008, 101, 388–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Görlich, D. Nuclear protein import. Curr. Opin. Cell Biol. 1997, 9, 412–419. [Google Scholar] [CrossRef] [Green Version]
- Hu, Q.; Paau, M.C.; Zhang, Y.; Chan, W.; Gong, X.; Zhang, L.; Choi, M.M.F. Capillary electrophoretic study of amine/carboxylic acid-functionalized carbon nanodots. J. Chromatogr. A 2013, 1304, 234–240. [Google Scholar] [CrossRef]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Ji, Z.; Zeng, Z.; Jayapalan, A.; Bagra, B.; Sheardy, A.; He, P.; LaJeunesse, D.R.; Wei, J. Dark-Field Microscopic Study of Cellular Uptake of Carbon Nanodots: Nuclear Penetrability. Molecules 2022, 27, 2437. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27082437
Zhang W, Ji Z, Zeng Z, Jayapalan A, Bagra B, Sheardy A, He P, LaJeunesse DR, Wei J. Dark-Field Microscopic Study of Cellular Uptake of Carbon Nanodots: Nuclear Penetrability. Molecules. 2022; 27(8):2437. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27082437
Chicago/Turabian StyleZhang, Wendi, Zuowei Ji, Zheng Zeng, Anitha Jayapalan, Bhawna Bagra, Alex Sheardy, Peng He, Dennis R. LaJeunesse, and Jianjun Wei. 2022. "Dark-Field Microscopic Study of Cellular Uptake of Carbon Nanodots: Nuclear Penetrability" Molecules 27, no. 8: 2437. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27082437