A QSAR Toxicity Study of a Series of Alkaloids with the Lycoctonine Skeleton
Abstract
:Introduction
Materials and Methods
Training Compounds

| Compound | R | R1 | R2 | R3 | R4 |
|---|---|---|---|---|---|
| Neoline | OH | CH2OCH3 | αOCH3 | H | OH |
| Isotalatisidine | OH | CH2OCH3 | H | H | OH |
| Кarakoline | OH | CH3 | H | H | OH |
| Delsoline | OH | CH2OCH3 | βOCH3 | OH | OCH3 |
| Kondelfine | OH | CH2OCH3 | H | H | OCOCH3 |
| 14-Acetylvirescenine | OH | CH2OCH3 | H | OH | OCOCH3 |
| Gigactonine | OH | CH2OH | βOCH3 | OH | OCH3 |
| Lycoctonine | OCH3 | CH2OH | βOCH3 | OH | OCH3 |
| Delectinine | OCH3 | CH2OH | βOCH3 | OH | OH |

| Compound | R | R1 | Compound | R | R1 |
|---|---|---|---|---|---|
| Lycaconitine | OCH3 |  | Delectine | OH | NH2 |
| Nudicauline | OCOCH3 |  | О- Acetyldelectine | OCOCH3 | NH2 |
| Меthyllycaconitine | OCH3 |  | N- Acetyldelectine | OH | NHCOCH3 |
| Anthranoyllycoctonine | OCH3 | NH2 | N-О-Diacetyldelectine | OCOCH3 | NHCOCH3 |
| Puberaconitine | OCH3 | NHCO(CH2)2COOH | Ajacine | OCH3 | NHCOCH3 |
Molecular Modelling
Molecular Descriptors
Statistical Methods
Results and Discussion
3D Descriptor Containing Models
| DiMo | lgP | E | HOMO | LUMO | GAP | MW | TPSA | MlogP | nHDon | nHAcc | nC=O | MR | Mor28e | RDF020u | Mor07p | |
| DiMo | 1 | .38 | .428 | .406 | .514 | .533 | .635 | .565 | .007 | .092 | .618 | .517 | .643 | .022 | .452 | .433 |
| lgP | 1 | .604 | .599 | .402 | .482 | .504 | .372 | .21 | .638 | .606 | .237 | .494 | .259 | .862 | .234 | |
| E | 1 | .487 | .554 | .607 | .784 | .761 | .044 | .228 | .882 | .669 | .737 | .035 | .594 | .571 | ||
| HOMO | 1 | .411 | .512 | .594 | .482 | .054 | .353 | .62 | .332 | .594 | .018 | .654 | .449 | |||
| LUMO | 1 | .985 | .857 | .811 | .118 | .092 | .792 | .788 | .884 | .034 | .397 | .728 | ||||
| GAP | 1 | .897 | .832 | .068 | .139 | .847 | .78 | .921 | .042 | .473 | .752 | |||||
| MW | 1 | .959 | .017 | .11 | .977 | .88 | .995 | .007 | .552 | .875 | ||||||
| TPSA | 1 | .044 | .035 | .93 | .952 | .941 | .001 | .4 | .909 | |||||||
| MlogP | 1 | .333 | 0 | .117 | .028 | .126 | .173 | .09 | ||||||||
| nHDon | 1 | .175 | .006 | .11 | .331 | .516 | .008 | |||||||||
| nHAcc | 1 | .833 | .958 | .021 | .622 | .8 | ||||||||||
| nC=O | 1 | .861 | .000 | .236 | .878 | |||||||||||
| MR | 1 | .006 | .555 | .87 | ||||||||||||
| Mor28e | 1 | .119 | .015 | |||||||||||||
| RDF020u | 1 | .283 | ||||||||||||||
| Mor07p | 1 |
| Alkaloid | LD50 mg/kg* |  | RDF020u | Mor28e | Mor07p | TPSA | nC=O | MlogP | MW |
| Neoline | 69.0 | 3.80 | 18.075 | -1.417 | 3.436 | 30.93 | 0 | 1.473 | 437.64 |
| Isotalatisidine | 40.1 | 4.00 | 16.387 | -1.184 | 3.438 | 21.7 | 0 | 2.021 | 407.61 |
| Кarakoline | 51.5 | 3.86 | 13.17 | -1.383 | 3.164 | 12.47 | 0 | 2.578 | 377.58 |
| Delsoline | 175.0 | 3.43 | 21.23 | -0.985 | 3.516 | 40.16 | 0 | 0.932 | 467.67 |
| Kondelfinee | 18.5 | 4.39 | 15.798 | -1.076 | 3.723 | 48 | 1 | 2.348 | 449.65 |
| 14-Acetylvirescenine | 18.0 | 4.41 | 16.249 | -0.84 | 3.749 | 48 | 1 | 1.599 | 465.65 |
| Lycaconotine | 2.6 | 5.41 | 20.806 | -1.257 | 6.025 | 103.84 | 3 | 1.673 | 668.86 |
| Nudicauline | 1.8 | 5.60 | 19.957 | -1.241 | 6.337 | 120.91 | 4 | 2.046 | 710.9 |
| Gigactonine | 88.0 | 3.71 | 19.164 | -0.754 | 3.463 | 30.93 | 0 | 0.725 | 453.64 |
| Lycoctonine | 170.0 | 3.44 | 20.081 | -1.149 | 3.458 | 40.16 | 0 | 0.932 | 467.67 |
| Delectinine | 130.0 | 3.54 | 20.173 | -0.903 | 3.228 | 30.93 | 0 | 0.725 | 453.64 |
| Methyllycaconitine | 3.9 | 5.24 | 21.178 | -1.236 | 6.248 | 103.84 | 3 | 1.85 | 682.89 |
| Anthranoyllycoctonine | 20.1 | 4.46 | 22.304 | -1.138 | 4.786 | 66.46 | 1 | 1.741 | 586.8 |
| Puberaconitine | 22.5 | 4.48 | 23.449 | -1.247 | 4.999 | 100.6 | 3 | 1.368 | 686.88 |
| Delectine | 35.8 | 4.20 | 22.1 | -0.898 | 4.25 | 57.23 | 1 | 1.553 | 572.77 |
| О-Acetyldelectine | 15.5 | 4.60 | 21.765 | -1.011 | 5.08 | 83.53 | 2 | 1.929 | 614.81 |
| N- Acetyldelectine | 25.3 | 4.39 | 22.521 | -0.641 | 4.522 | 74.3 | 2 | 1.523 | 614.81 |
| N-О-Diacetyldelectine | 12.5 | 4.71 | 21.3 | -0.833 | 4.98 | 100.6 | 3 | 1.9 | 656.85 |
| Ajacine | 9.0 | 4.84 | 22.423 | -0.683 | 4.733 | 83.53 | 2 | 1.706 | 628.84 |



Physicochemical descriptor containing models.

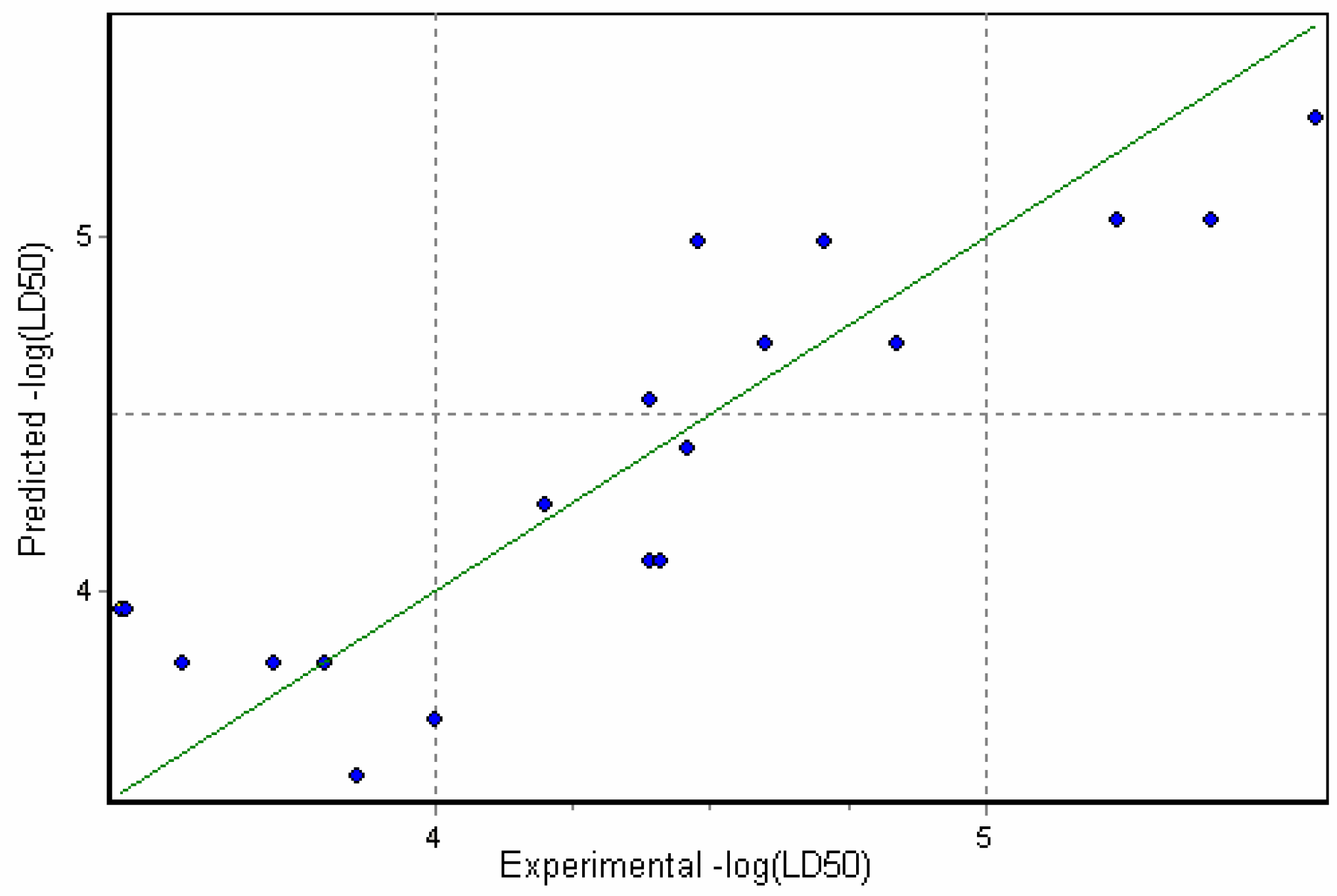
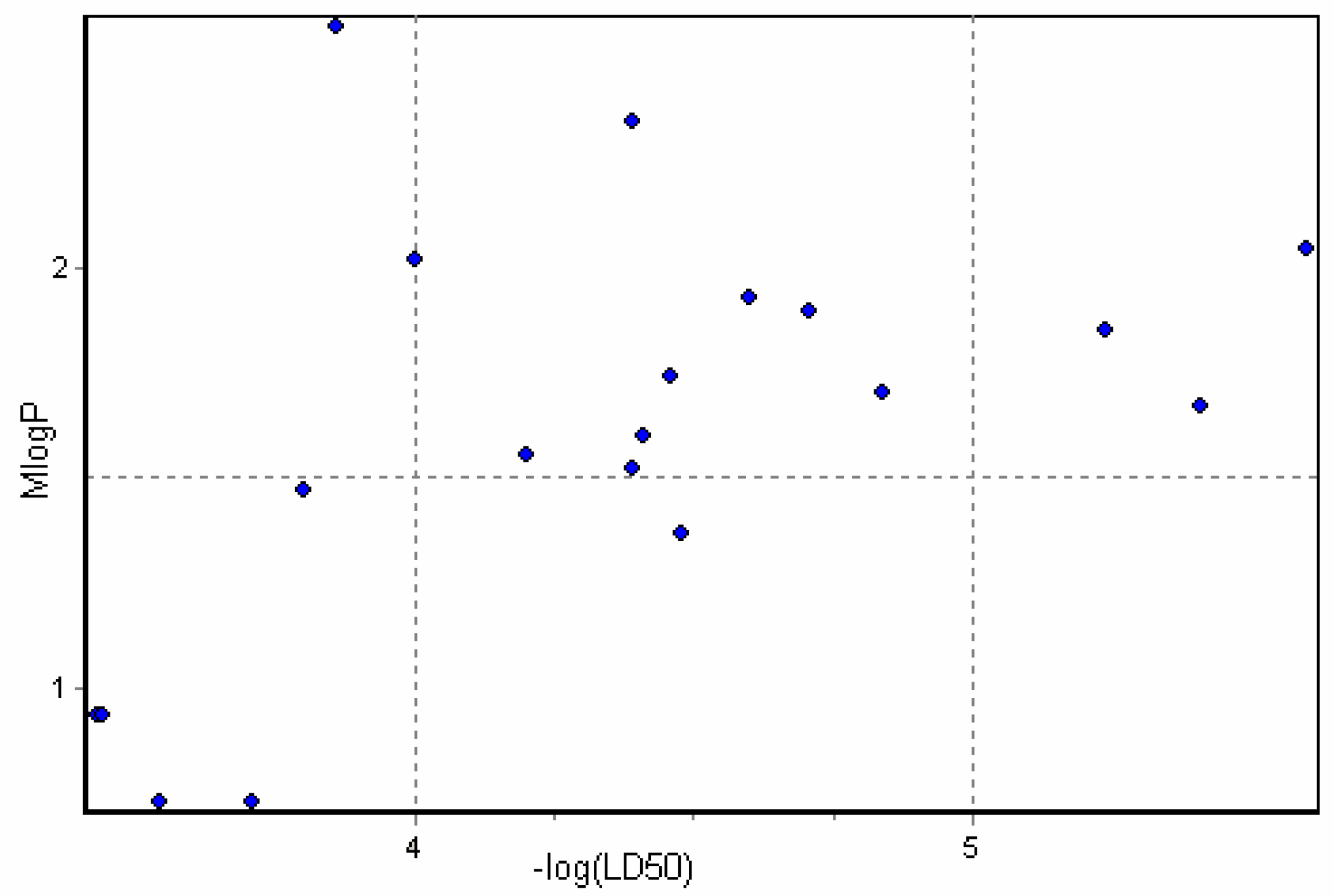

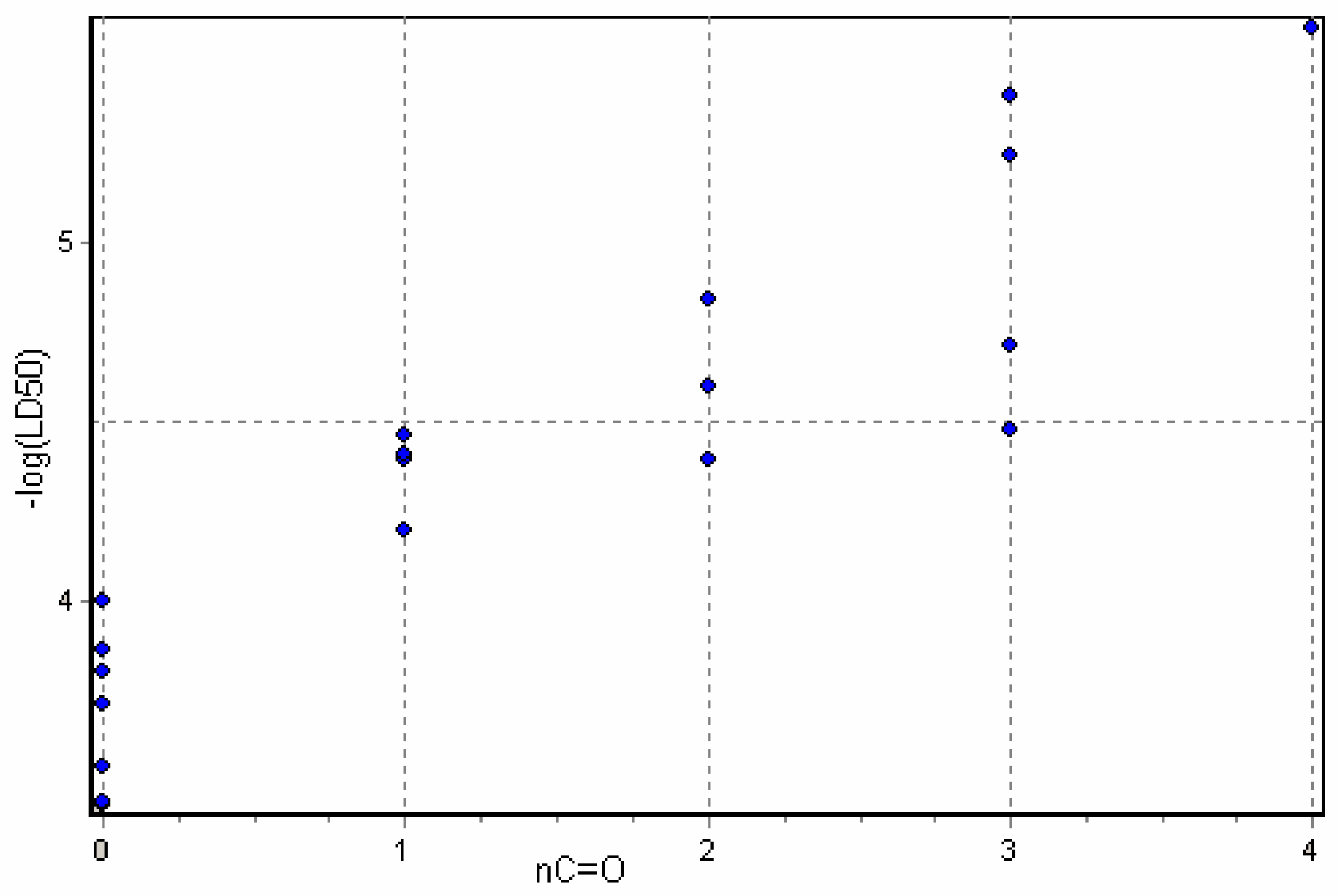



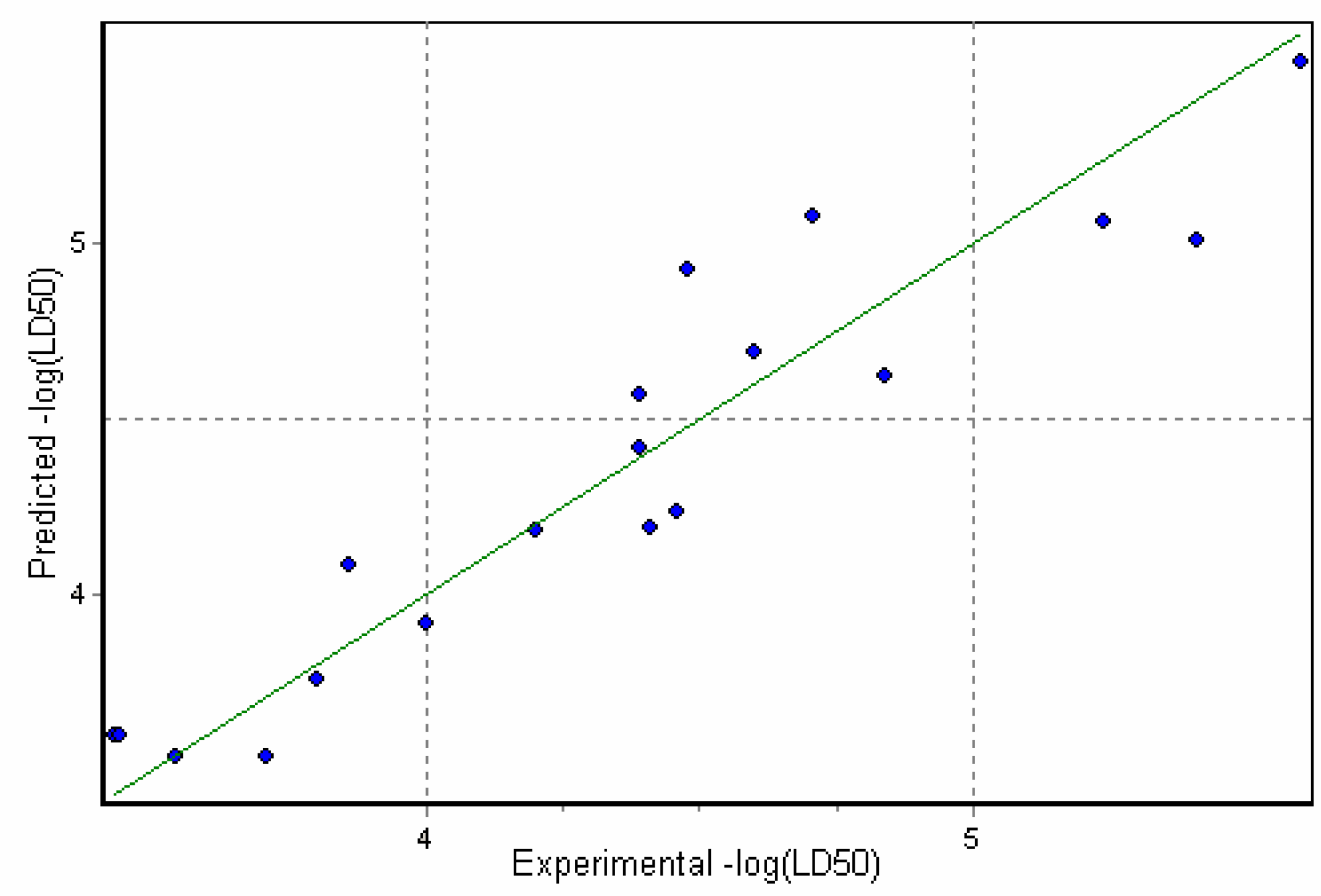

Conclusions
Acknowledgments
References and Notes
- Ameri, A. Prog. Neurobiol. 1998, 56, 211.
- Dzhakhangirov, F. N.; Sultankhodzhaev, M. N.; Tashkhodzhaev, B.; Salimov, B. T. Chem. Nat. Comp. 1997, 33, 190. [CrossRef]
- Zhu, D. Y.; Bai, D.L.; Tang, X.C. Drug Dev. Res. 1996, 39, 147.
- Heubach, J.F.; Schule, A. Planta Med. 1998, 64, 22.
- Panter, K.E.; Manners, G.D.; Stegelmeier, B.L.; Lee, S.; Gardner, D.R.; Ralphs, M.H.; Pfister, J.A.; James, L.F. Biochem. Syst. Ecol. 2002, 30, 113. [CrossRef]
- Siemion, R.S.; Raisbek, M.F.; Waggoner, J.W.; Tidwell, M.A.; Sanchez, D.A. Vet. Human Toxicol. 1992, 34, 206.
- Bello-Ramirez, A.M.; Buendia-Orozco, J.; Nava-Ocampo, A.A. Fundam. Clin. Pharmacol. 2003, 17, 575.
- Fujita, T. QSAR and Drug Design; New Developments and Applications; Elsevier: Amsterdam, (The Netherlands), 1995. [Google Scholar]
- Lee, D. L.; Kollman, P. A.; Marsh, F. J.; Wolff, M. E. J. Med. Chem. 1977, 20, 1139. [CrossRef]
- Kubinyi, H. 3D QSAR in Drug Design: Theory, Methods and Applications; ESCOM: Amsterdam, (The Netherlands), 1993. [Google Scholar]
- Zhou, Y.-X.; Xu, L.; Wu, Y.-P.; Liu, B.-L. J. Chem. Intell. Lab. Sys. 1999, 45, 95. [CrossRef]
- Hou, T. J.; Wang, J. M.; Liao, N.; Xu, X. J. J. Chem. Inf. Comput. Sci. 1999, 39, 775. [CrossRef]
- Davis, L. Handbook of Genetic Algorithms; Van Nostrand Reinhold: New York (USA), 1991. [Google Scholar]
- Devillers, J. Genetic Algorithms in Molecular Modeling; Academic Press Ltd.: London, (U.K.), 1996. [Google Scholar]
- Dzhakhangirov, F. N.; Salimov, B. T.; Bessonova, I. A.; Sultankhodzhaev, M. N. Chem. Nat.Comp. 1995, 31, 708. [CrossRef]
- HyperChem 6.01 (HyperCube, Inc), 2000
- Young, D. Computational Chemistry: A Practical Guide for Applying Techniques to Real World Problems; John-Wiley & Sons: New York (USA), 2001. [Google Scholar]
- Todeschini, R.; Consonni, V. DRAGON software for the Calculation of Molecular Descriptors, Web version 3.0 for Windows.
- Todeschini, R.; Consonni, V. Handbook of Molecular Descriptors; Mannhold, R., Kubinyi, H., Timmerman, H., Eds.; Wiley-VCH: Weinheim, New York, 2000. [Google Scholar]
- de Oliveira, D.B.; Gaudio, A.C. Quant. Struct.-Act. Relat. 2000, 19, 599.
- Statistical analysis software NCSS98, trial version, 1998
- Cronin, M.T.D.; Schultz, T. W. J. Mol. Struct. (Theochem) 2003, 622, 39. [CrossRef]
- Schuur, J.H.; Selzer, P.; Gasteiger, J. J. Chem. Inf. Comput. Sci. 1996, 36, 334. [CrossRef]
- Hemmer, M.C.; Steinhauer, V.; Gasteiger, J. Vibrat. Spectr. 1999, 19, 151.
- Ertl, P.; Rohde, B.; Selzer, P. J. Med. Chem. 2000, 43, 3714. [CrossRef]
- Kelder, J.; Grootenhuis, P.D.J.; Bayada, D.M.; Delbressine, L.P.C.; Ploemen, J-P. Pharm. Res. 1999, 16, 1514. [CrossRef] [PubMed]
- Clark, D.E. J. Pharm. Sci. 1999, 88, 815. [CrossRef]
- Clark, D.E. J. Pharm. Sci. 1999, 88, 807. [CrossRef]
- Aronov, A. M.; Goldman, B. B. Bioorg. Med. Chem. 2004, 12, 2307.
- Kutzenko, S.A. Osnovy Toxikologii (Fundamentals of Toxicology) Sankt-Petersburg. 2002. (electronic version at www.medline.ru/monograf/toxicology).
© 2004 by MDPI (http://www.mdpi.org). Reproduction is permitted for noncommercial purposes.
Share and Cite
Turabekova, M.A.; Rasulev, B.F. A QSAR Toxicity Study of a Series of Alkaloids with the Lycoctonine Skeleton. Molecules 2004, 9, 1194-1207. https://0-doi-org.brum.beds.ac.uk/10.3390/91201194
Turabekova MA, Rasulev BF. A QSAR Toxicity Study of a Series of Alkaloids with the Lycoctonine Skeleton. Molecules. 2004; 9(12):1194-1207. https://0-doi-org.brum.beds.ac.uk/10.3390/91201194
Chicago/Turabian StyleTurabekova, Malakhat A., and Bakhtiyor F. Rasulev. 2004. "A QSAR Toxicity Study of a Series of Alkaloids with the Lycoctonine Skeleton" Molecules 9, no. 12: 1194-1207. https://0-doi-org.brum.beds.ac.uk/10.3390/91201194




