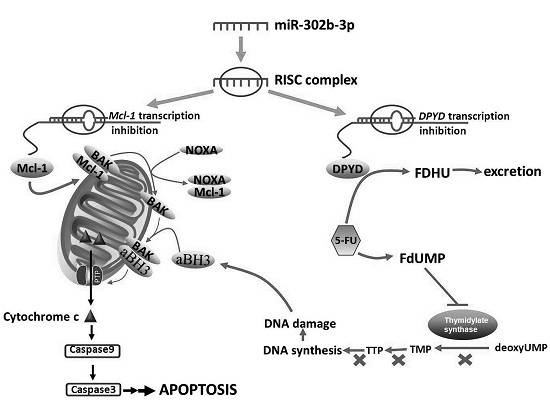
MicroRNA-302b Enhances the Sensitivity of Hepatocellular Carcinoma Cell Lines to 5-FU via Targeting Mcl-1 and DPYD
Abstract
:1. Introduction
2. Results and Discussion
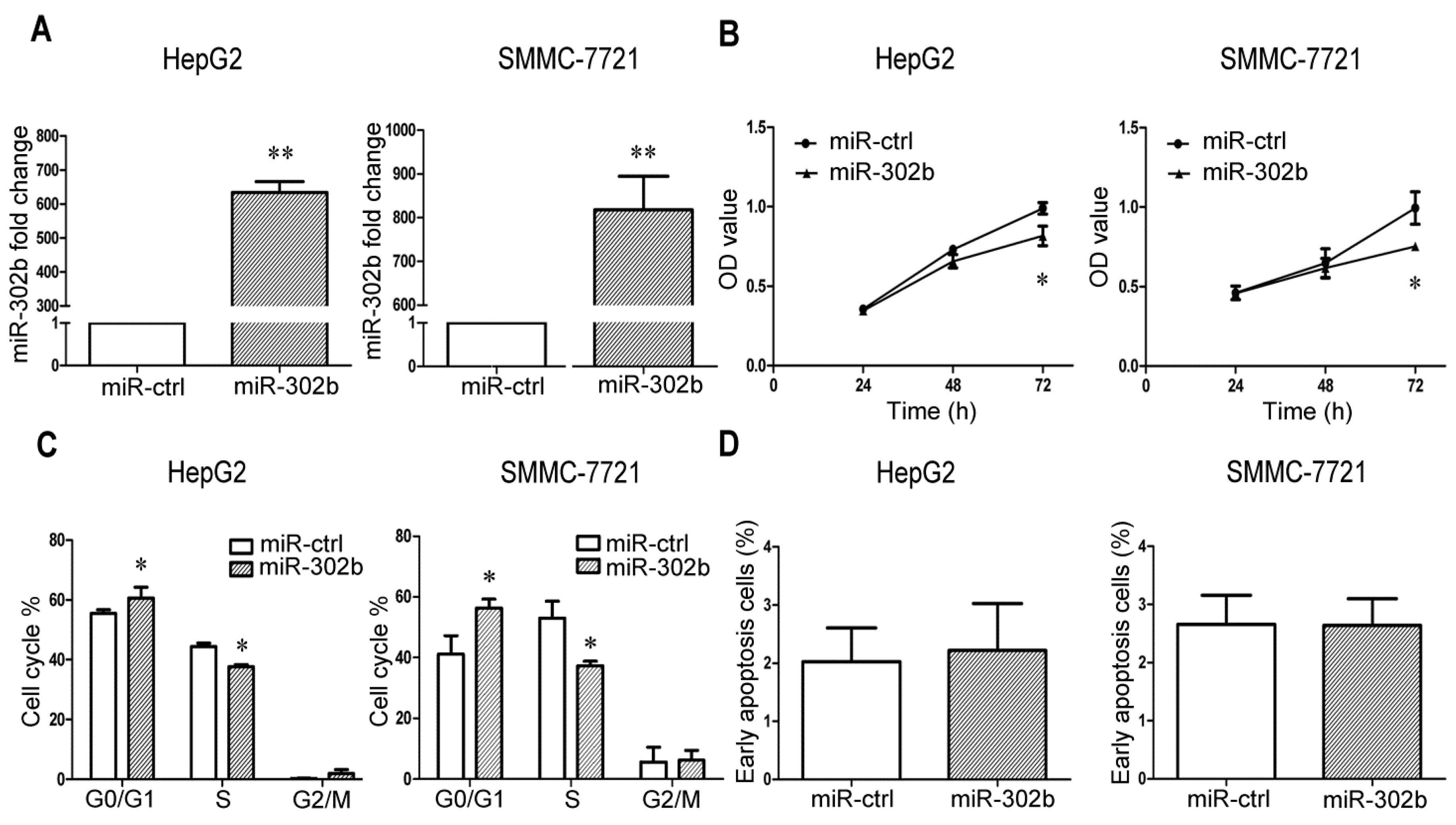
2.1. MiR-302b Represses Cell Proliferation and Cell Cycle Progression in Liver Cancer Cell Lines
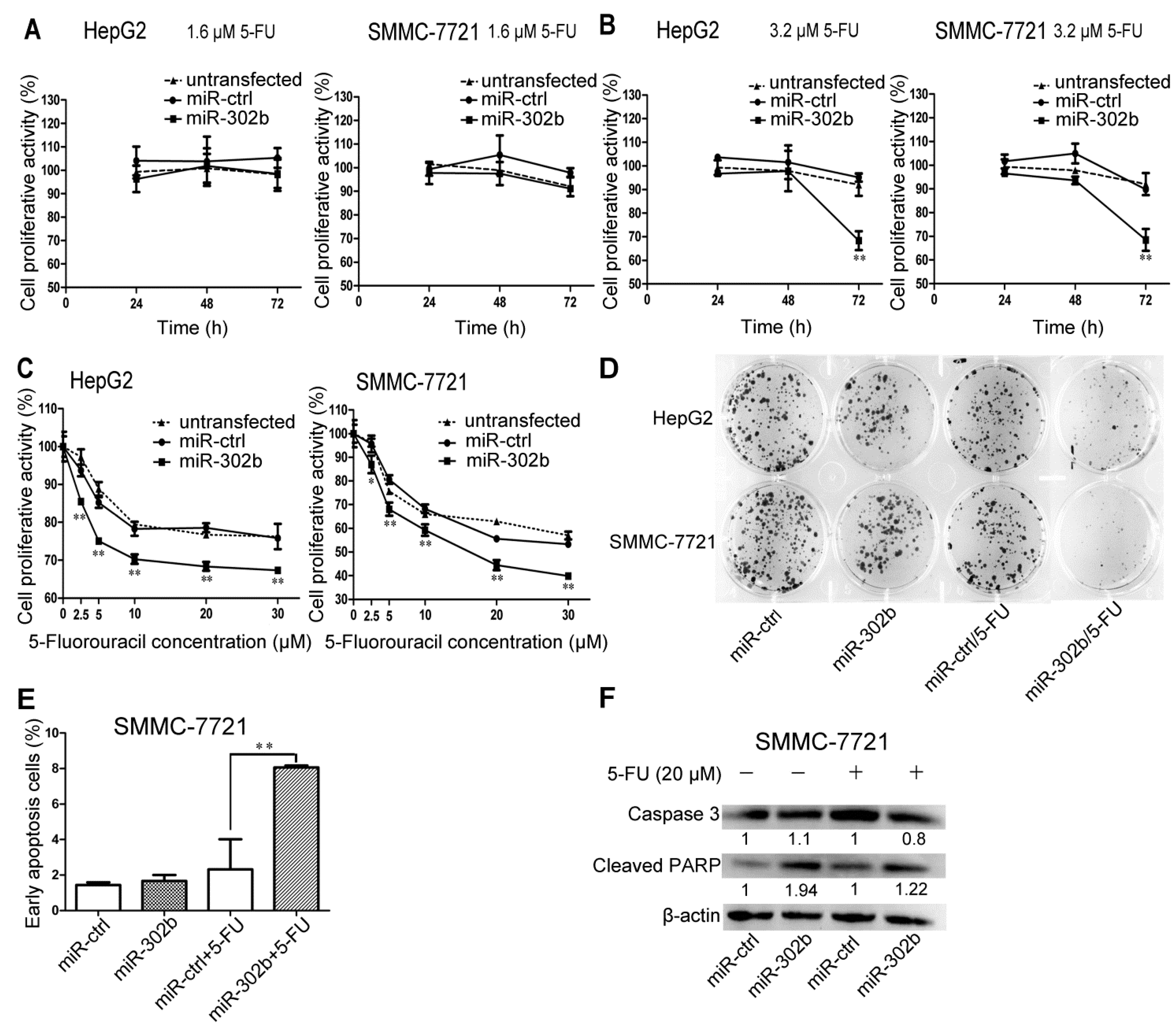
2.2. Overexpression of miR-302b Enhances the Sensitivity to 5-FU in HepG2 and SMMC-7721 Cells
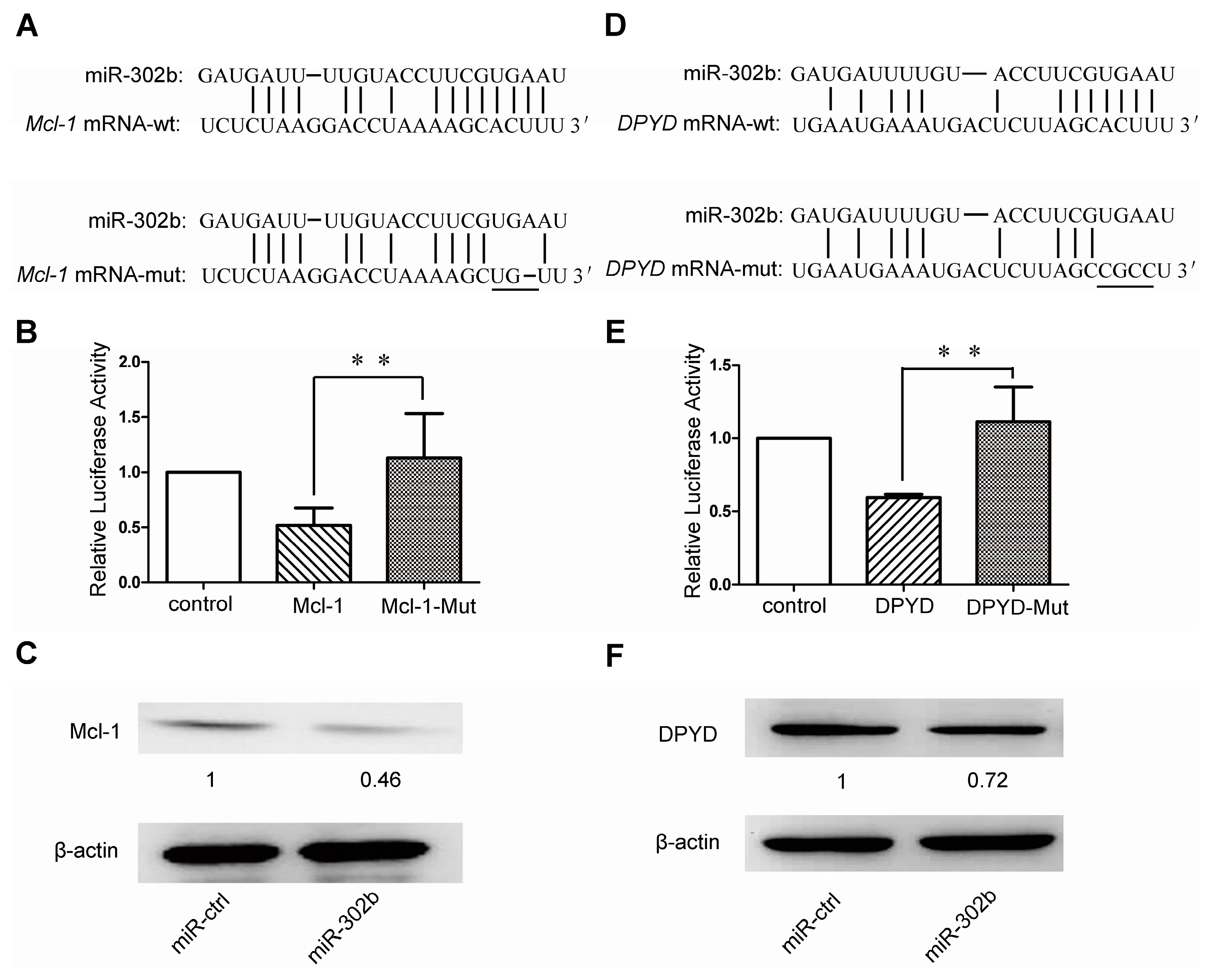
2.3. MiR-302b Targets Mcl-1 and DPYD
2.4. RNA Interference-Mediated Silencing of Mcl-1 or DPYD Enhances the Sensitivity to 5-FU in HepG2 and SMMC-7721 Cells

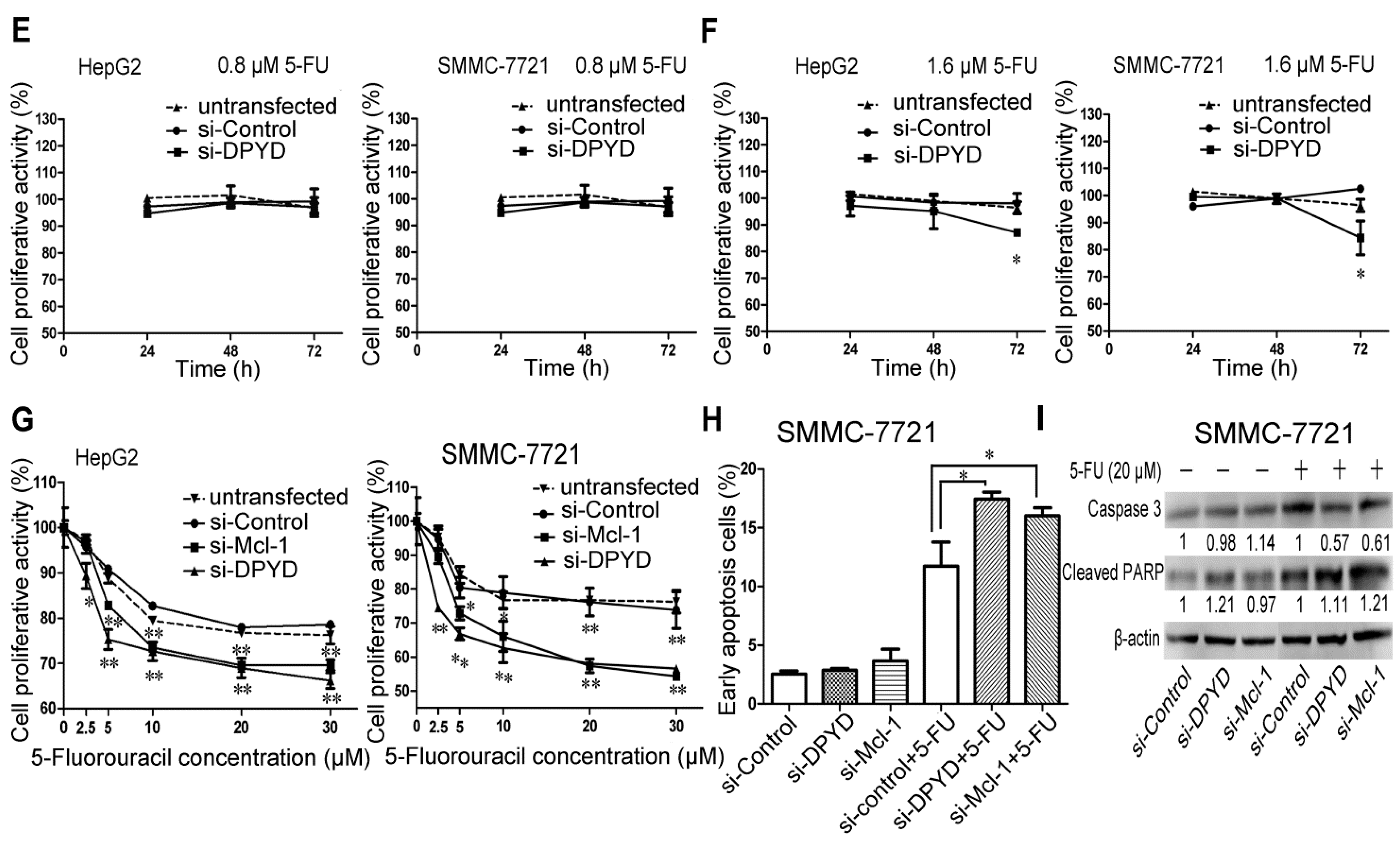
2.5. Transfection of miR-302b Sensitizes the HCC Cell Lines to Apoptosis Induced by 5-FU
3. Experimental Section
3.1. Cell Cultures and Chemicals
3.2. Plasmid Constructions
3.3. MiRA/siRNA Transfection
3.4. qRT-PCR Assay
| Name | Sequence 5′–3′ |
|---|---|
| Pre-miR-302b-F | AATTCGCTCCCTTCAACTTTAACATGGAAGTGCTTTCTGTGACTTTAAAAGTAAGTGCTTCCATGTTTTAGTAGGAGTA |
| Pre-miR-302b-R | AGCTTACTCCTACTAAAACATGGAAGCACTTACTTTTAAAGTCACAGAAAGCACTTCCATGTTAAAGTTGAAGGGAGCG |
| Mcl-1-UTR-F | TCGAGGTATCTCTAAGGACCTAAAAGCACTTTATGG |
| Mcl-1-UTR-R | TCGACCATAAAGTGCTTTTAGGTCCTTAGAGATACC |
| Mcl-1-UTR-MF | TCGAGGTATCTCTAAGGACCTAAAAGCTGTTATGG |
| Mcl-1-UTR-MR | TCGACCATAACAGCTTTTAGGTCCTTAGAGATACC |
| DPYD-CDS-F | TCGAGTGAATGAAATGACTCTTAGCACTTTG |
| DPYD-CDS-R | TCGACAAAGTGCTAAGAGTCATTTCATTCAC |
| DPYD-CDS-MF | TCGAGTGAATGAAATGACTCTTAGCCGCCTG |
| DPYD-CDS-MR | TCGACAGGCGGCTAAGAGTCATTTCATTCAC |
| miR-302b-RT | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCTACTA |
| miR-302b-F | TGCTTAAGTGCTTCCATGTT |
| miR-302b-R | ATCCAGTGCGTGTCGTG |
| β-actin-F | CCAACCGCGAGAAGATGA |
| β-actin-R | CCAGAGGCGTACAGGGATAG |
| Mcl-1L-F | GGAGGAGGAGGACGAGTTGTA |
| Mcl-1L-R | TGATGTCCAGTTTCCGAAGCA |
| Mcl-1S-F | GAGACGGCCTTCCAAGGAT |
| Mcl-1S-R | AGGTTGCTAGGGTGCAACTCT |
| DPYD-F | GAACTTGCCAAGAAGTCTGAGG |
| DPYD-R | ATGCCATGTGGACATGATAAAT |
| U6-RT | AACGCTTCACGAATTTGCGT |
| U6-F | CTCGCTTCGGCAGCACA |
| U6-R | AACGCTTCACGAATTTGCGT |
| si-Mcl-1-F | GGACUUUUAGAUUUAGUGAUU |
| si-Mcl-1-R | UCACUAAAUCUAAAAGUCCUU |
| si-DPYD-F | UAUUGUAACUGCACAUAAUGCUAGCUU |
| si-DPYD-R | GCUAGCAUUAUGUGCAGUUACAAUAUU |
3.5. Cell Viability Assay
3.6. Luciferase Assay
3.7. Cell Cycle Assays by Flow Cytometry
3.8. 5-FU Sensitivity Assay
3.9. Colony Assay
3.10. Cell Apoptosis Assay by Flow Cytometry
3.11. Western Blotting
3.12. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Conflicts of Interest
References
- Chen, W.; Zheng, R.; Zeng, H.; Zhang, S.; He, J. Annual report on status of cancer in china, 2011. Chin. J. Cancer Res. 2015, 27. [Google Scholar] [CrossRef]
- Gouill, S.L.; Podar, K.; Harousseau, J.L.; Anderson, K.C. Mcl-1 regulation and its role in multiple myeloma. Cell Cycle 2004, 3, 1259–1262. [Google Scholar] [CrossRef] [PubMed]
- Fleischer, B.; Schulze-Bergkamen, H.; Schuchmann, M.; Weber, A.; Biesterfeld, S.; Müller, M.; Krammer, P.H.; Galle, P.R. Mcl-1 is an anti-apoptotic factor for human hepatocellular carcinoma. Int. J. Oncol. 2006, 28, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Coppola, D.; Livingston, S.; Cress, W.D.; Haura, E.B. Mcl-1 regulates survival and sensitivity to diverse apoptotic stimuli in human non-small cell lung cancer cells. Cancer Biol. Ther. 2005, 4, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Nijhawan, D.; Fang, M.; Traer, E.; Zhong, Q.; Gao, W.; Du, F.; Wang, X. Elimination of Mcl-1 is required for the initiation of apoptosis following ultraviolet irradiation. Genes Dev. 2003, 17, 1475–1486. [Google Scholar] [CrossRef] [PubMed]
- Wertz, I.E.; Kusam, S.; Lam, C.; Okamoto, T.; Sandoval, W.; Anderson, D.J.; Helgason, E.; Ernst, J.A.; Eby, M.; Liu, J. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature 2011, 471, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Schulze-Bergkamen, H.; Fleischer, B.; Schuchmann, M.; Weber, A.; Weinmann, A.; Krammer, P.H.; Galle, P.R. Suppression of Mcl-1 via RNA interference sensitizes human hepatocellular carcinoma cells towards apoptosis induction. BMC Cancer 2006, 6. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Gong, W.H.; Li, X.C.; Zou, C.P.; Jiang, G.J.; Li, X.H.; Qian, H. Oxaliplatin sensitizes OS cells to trail-induced apoptosis via down-regulation of Mcl-1. Asian Pac. J. Cancer Prev. 2012, 13, 3477–3481. [Google Scholar] [CrossRef] [PubMed]
- Naguib, F.N.; el Kouni, M.H.; Cha, S. Enzymes of uracil catabolism in normal and neoplastic human tissues. Cancer Res. 1985, 45, 5405–5412. [Google Scholar] [PubMed]
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Fukui, Y.; Oka, T.; Nagayama, S.; Danenberg, P.V.; Danenberg, K.D.; Fukushima, M. Thymidylate synthase, dihydropyrimidine dehydrogenase, orotate phosphoribosyltransferase mRNA and protein expression levels in solid tumors in large scale population analysis. Int. J. Mol. Med. 2008, 22, 709–716. [Google Scholar] [PubMed]
- Noguchi, T.; Tanimoto, K.; Shimokuni, T.; Ukon, K.; Tsujimoto, H.; Fukushima, M.; Noguchi, T.; Kawahara, K.; Hiyama, K.; Nishiyama, M. Aberrant methylation of DPYD promoter, DPYD expression, and cellular sensitivity to 5-fluorouracil in cancer cells. Clin. Cancer Res. 2004, 10, 7100–7107. [Google Scholar] [CrossRef] [PubMed]
- Yoo, B.K.; Gredler, R.; Vozhilla, N.; Su, Z.Z.; Chen, D.; Forcier, T.; Shah, K.; Saxena, U.; Hansen, U.; Fisher, P.B. Identification of genes conferring resistance to 5-fluorouracil. Proc. Natl. Acad. Sci. USA 2009, 106, 12938–12943. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Wiemer, E.A. The role of microRNAs in cancer: No small matter. Eur. J. Cancer 2007, 43, 1529–1544. [Google Scholar] [CrossRef] [PubMed]
- Wienholds, E.; Plasterk, R.H. MicroRNA function in animal development. FEBS Lett. 2005, 579, 5911–5922. [Google Scholar] [CrossRef] [PubMed]
- Drakaki, A.; Hatziapostolou, M.; Iliopoulos, D. Therapeutically targeting microRNAs in liver cancer. Curr. Pharm. Des. 2013, 19, 1180–1191. [Google Scholar] [PubMed]
- Gotanda, K.; Hirota, T.; Matsumoto, N.; Ieiri, I. MicroRNA-433 negatively regulates the expression of thymidylate synthase (TYMS) responsible for 5-fluorouracil sensitivity in hela cells. BMC Cancer 2013, 13. [Google Scholar] [CrossRef] [PubMed]
- Puca, A.A.; Daly, M.J.; Brewster, S.J.; Matise, T.C.; Barrett, J.; Shea-Drinkwater, M.; Kang, S.; Joyce, E.; Nicoli, J.; Benson, E. A genome-wide scan for linkage to human exceptional longevity identifies a locus on chromosome 4. Proc. Natl. Acad. Sci. USA 2001, 98, 10505–10508. [Google Scholar] [CrossRef] [PubMed]
- Suh, M.R.; Lee, Y.; Kim, J.Y.; Kim, S.K.; Moon, S.H.; Lee, J.Y.; Cha, K.Y.; Chung, H.M.; Yoon, H.S.; Moon, S.Y. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 2004, 270, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Barroso-del Jesus, A.; Lucena-Aguilar, G.; Menendez, P. The miR-302-367 cluster as a potential stemness regulator in ESCs. Cell Cycle 2009, 8, 394–398. [Google Scholar] [CrossRef] [PubMed]
- Rosa, A.; Brivanlou, A.H. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J. 2011, 30, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.L.; Chang, D.C.; Lin, C.H.; Ying, S.Y.; Leu, D.; Wu, D.T. Regulation of somatic cell reprogramming through inducible miR-302 expression. Nucleic Acids Res. 2011, 39, 1054–1065. [Google Scholar] [CrossRef] [PubMed]
- Subramanyam, D.; Lamouille, S.; Judson, R.L.; Liu, J.Y.; Bucay, N.; Derynck, R.; Blelloch, R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat. Biotechnol. 2011, 29, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.L.; Chang, D.C.; Ying, S.Y.; Leu, D.; Wu, D.T. MicroRNA miR-302 inhibits the tumorigenecity of human pluripotent stem cells by coordinate suppression of the CDK2 and CDK4/6 cell cycle pathways. Cancer Res. 2010, 70, 9473–9482. [Google Scholar] [CrossRef] [PubMed]
- Cai, N.; Wang, Y.D.; Zheng, P.S. The microRNA-302-367 cluster suppresses the proliferation of cervical carcinoma cells through the novel target AKT1. RNA 2013, 19, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yao, J.; Zhang, X.; Guo, B.; Le, X.; Cubberly, M.; Li, Z.; Nan, K.; Song, T.; Huang, C. MiRNA-302b suppresses human hepatocellular carcinoma by targeting AKT2. Mol. Cancer Res. 2014, 12, 190–202. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.H.; Huang, H.Y.; Hsu, J.B.; Weng, S.L.; Horng, J.T.; Huang, H.D. An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinform. 2013, 14. [Google Scholar] [CrossRef]
- Coronnello, C.; Benos, P.V. Comir: Combinatorial microRNA target prediction tool. Nucleic Acids Res. 2013, 41, W159–W164. [Google Scholar] [CrossRef] [PubMed]
- Koga, C.; Kobayashi, S.; Nagano, H.; Tomimaru, Y.; Hama, N.; Wada, H.; Kawamoto, K.; Eguchi, H.; Konno, M.; Ishii, H. Reprogramming using microRNA-302 improves drug sensitivity in hepatocellular carcinoma cells. Ann. Surg. Oncol. 2014, 21, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Jopling, C.L.; Yi, M.; Lancaster, A.M.; Lemon, S.M.; Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 2005, 309, 1577–1581. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Lin, H.; Pan, Z.; Xiao, J.; Zhang, Y.; Lu, Y.; Yang, B.; Wang, Z. Down-regulation of miR-1/miR-133 contributes to re-expression of pacemaker channel genes HCN2 and HCN4 in hypertrophic heart. J. Biol. Chem. 2008, 283, 20045–20052. [Google Scholar] [CrossRef] [PubMed]
- Tay, Y.; Zhang, J.; Thomson, A.M.; Lim, B.; Rigoutsos, I. MicroRNAs to Nanog, OCT4 and SOX2 coding regions modulate embryonic stem cell differentiation. Nature 2008, 455, 1124–1128. [Google Scholar] [CrossRef] [PubMed]
- Adams, K.W.; Cooper, G.M. Rapid turnover of Mcl-1 couples translation to cell survival and apoptosis. J. Biol. Chem. 2007, 282, 6192–6200. [Google Scholar] [CrossRef] [PubMed]
- Someya, M.; Sakata, K.I.; Matsumoto, Y.; Tauchi, H.; Kai, M.; Hareyama, M.; Fukushima, M. Effects of depletion of dihydropyrimidine dehydrogenase on focus formation and RPA phosphorylation. J. Radiat. Res. 2012, 53, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.; Leo, C.P.; Hsu, S.Y.; Hsueh, A.J. MCL-1S, a splicing variant of the antiapoptotic BCL-2 family member MCL-1, encodes a proapoptotic protein possessing only the BH3 domain. J. Biol. Chem. 2000, 275, 25255–25261. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, D.; He, K.; Chang, S.; Tong, D.; Huang, C. MicroRNA-302b Enhances the Sensitivity of Hepatocellular Carcinoma Cell Lines to 5-FU via Targeting Mcl-1 and DPYD. Int. J. Mol. Sci. 2015, 16, 23668-23682. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms161023668
Cai D, He K, Chang S, Tong D, Huang C. MicroRNA-302b Enhances the Sensitivity of Hepatocellular Carcinoma Cell Lines to 5-FU via Targeting Mcl-1 and DPYD. International Journal of Molecular Sciences. 2015; 16(10):23668-23682. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms161023668
Chicago/Turabian StyleCai, Donghui, Kang He, Su'e Chang, Dongdong Tong, and Chen Huang. 2015. "MicroRNA-302b Enhances the Sensitivity of Hepatocellular Carcinoma Cell Lines to 5-FU via Targeting Mcl-1 and DPYD" International Journal of Molecular Sciences 16, no. 10: 23668-23682. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms161023668