Differentially-Expressed Genes Associated with Faster Growth of the Pacific Abalone, Haliotis discus hannai
Abstract
:1. Introduction
2. Results and Discussion
2.1. Differential Growth in Pacific Abalones
| Groups | Shell Length (mm) | Shell Width (mm) | Shell Height (mm) | Weight (g) |
|---|---|---|---|---|
| Small (n = 10) | 13.53 ± 1.34 | 9.54 ± 1.11 | 2.87 ± 0.41 | 0.26 ± 0.09 |
| Medium (n = 10) | 24.12 ± 2.49 | 16.35 ± 1.45 | 4.95 ± 0.46 | 1.43 ± 0.40 |
| Large (n = 10) | 36.15 ± 2.21 | 24.99 ± 1.43 | 8.86 ± 1.19 | 5.24 ± 1.09 |
| Mean ± SD (n = 80) | 24.63 ± 8.17 | 17.09 ± 5.58 | 5.54 ± 2.26 | 2.10 ± 1.81 |
2.2. Transcriptome Analysis
| Comparison (Total: RPKM ≥ 0.3) | FC ≥ 2, FDR ≤ 0.05 | FC ≥ 4, FDR ≤ 0.01 | FC ≥ 10, FDR ≤ 0.001 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Up | Down | Total | Up | Down | Total | Up | Down | Total | ||
| Large vs. Medium | 22,680 | 4383 | 4842 | 9225 | 1301 | 908 | 2209 | 190 | 141 | 331 |
| Large vs. Small | 24,414 | 10,957 | 4650 | 15,607 | 2859 | 1545 | 4404 | 543 | 428 | 971 |
| Medium vs. Small | 22,338 | 16,406 | 5249 | 21,655 | 4216 | 1907 | 6123 | 812 | 514 | 1326 |
2.3. Reverse Transcription-PCR (RT-PCR) Analysis
| Gene ID | RPKM | E Value* | Description | bp | Primer 5′–3′ | ||
|---|---|---|---|---|---|---|---|
| Large | Medium | Small | |||||
| Tsc1 | 1294.33 | 1227.59 | 1392.61 | 0 | Ribosomal protein L3 (D1M828) | 1401 | F: 5′-TGTCACCATCCTTGAGGCAC |
| R: 5′-CAGGAACAGGCTTCTCCAGG | |||||||
| Tsc2 | 10.03 | 4.08 | 0.88 | 5.00 × 10−16 | Incilarin A (B6RB57) | 709 | F: 5′-GGCGGCTACCTGGTTGAAAT |
| R: 5′-CCCACGATGTCTGGTCCATC | |||||||
| Tsc3 | 0.7 | 0.19 | 0.04 | 2.00 × 10−18 | Multiple epidermal growth factor-like domains 6 | 451 | F: 5′-GTACTGCCAGCTGGGTTCAT |
| R: 5′-CACCCGCCATTACAGCCAT | |||||||
| Tsc4 | 0.08 | 0.99 | 0 | 2.00 × 10−23 | Multiple epidermal growth factor-like domains 11 | 488 | F: 5′-CGGGTGGTTTGGAGAAGAGT |
| R: 5′-AGGCATGATCCACAATGCGA | |||||||
| Tsc5 | 19.26 | 11.71 | 2.74 | 7.00 × 10−52 | Perlucin (P82596) | 375 | F: 5′-CCTCTTGGGTTTATGCAGCAC |
| R: 5′-CGGACTGTCTCATTTCCAGAC | |||||||
| Tsc6 | 8.59 | 6.11 | 1.86 | 5.00 × 10−15 | EGF-like domain containing protein | 1222 | F: 5′-CACGGACATCACTGCAATAAGC |
| R: 5′-GACATGGCTCATGCGTCTCA | |||||||
| Tsc7 | 4.08 | 1.55 | 0.95 | 2.00 × 10−21 | Multiple epidermal growth factor-like domains 10 | 529 | F: 5′-GCAAGCACTGCAACCAAACT |
| R: 5′-TGCAACTTCTCTCCAGGCAG | |||||||
| Tsc8 | 33.68 | 23.4 | 7.37 | 3.00 × 10−8 | Transforming growth factor-β-induced protein ig-h3 (K1PUI7) | 399 | F: 5′-GTACGCATGGTCACCTACCC |
| R: 5′-TCGTAGCAGGGGTATGTTGTTA | |||||||
| Tsc9 | 2.27 | 1.14 | 0.43 | 4.00 × 10−18 | EGF-like domain containing protein | 745 | F: 5′-CTGACTGCAAGAGAGGCTGG |
| R: 5′-CTTGACATCCCCCTGGACAC | |||||||
| Tsc10 | 0.54 | 0.42 | 0.16 | 1.00 × 10−38 | Insulin-like growth factor-binding protein | 1123 | F: 5′-AGAGGTTCCTCCTGCCATCA |
| R: 5′-GCTTGTAGAGGCACCAGTGT | |||||||
| Tsc11 | 2.71 | 5.29 | 0.15 | 2.00 × 10−51 | Peroxidasin-like protein | 938 | F: 5′-ACAGTCTCGATCTGATGGCTT |
| R: 5′-CCAGAATCTGTCGCCCACTT | |||||||
| Tsc12 | 37.27 | 24.87 | 7.63 | 7.00 × 10−11 | C-type lectin 4 | 430 | F: 5′-CCTCGGGCTCTGCGACTC |
| R: 5′-ACGCCAGTCACCCTCAGTTG | |||||||
| Tsc13 | 19.42 | 21.52 | 4.74 | 2.00 × 10−30 | Choline transporter-like protein 1 | 289 | F: 5′-CGTGGCAGCGATTAACTGTG |
| R: 5′-GCTCATGTAGTAAGGCCGCT | |||||||
| Tsc14 | 8.14 | 9.34 | 1.86 | 5.00 × 10−79 | Fibroblast growth factor receptor 2 | 1807 | F: 5′-TCTCTGCGTCGCCATTCTTT |
| R: 5′-CAGTTGGACCAGCGGAGAAT | |||||||
| Tsc15 | 2683.36 | 2020.81 | 979.78 | 1.00 × 10−36 | Dermatopontin 3 | 604 | F: 5′-TCCCTACTGCGTTTCTGCTG |
| R: 5′-CTACCTTGCAGGGAGCAACA | |||||||
| Tsc16 | 49.8 | 17.74 | 16.35 | 3.00 × 10−16 | Perlucin A2 protein | 658 | F: 5′-TCCACTGGGATTTGTGGAACA |
| R: 5′-GTTGGCCCCTTCTCCAGTTG | |||||||
| Tsc17 | 36.31 | 46.58 | 9.19 | 2.00 × 10−16 | CD63 antigen | 1413 | F: 5′-TGGATTCAGCAAACCCGGAA |
| R: 5′-CAGGAAGCTTTGGGGTCGAA | |||||||
| Tsc18 | 329.2 | 276.28 | 216.62 | 2.00 × 10−144 | Cathepsin L2 cysteine protease; EC | 1090 | F: 5′-GTCGACTGGAGAACCAAGGG |
| R: 5′-GACAGAGATGGGACCAACGG | |||||||
| Tsc19 | 212.38 | 144.81 | 1636.37 | 0.00000007 | Calcium binding protein 1 | 457 | F: 5′-GGTGATAACCGTCTGACCGA |
| R: 5′-GGTGACAAATTCGCTTCGGG | |||||||
| Tsc20 | 45.46 | 25.82 | 374.98 | 1.00 × 10−23 | Tomoregulin-2 (K1Q9X6) | 722 | F: 5′-ACTGTGACCATGAGCGTGAG |
| R: 5′-CAGCTACGTGGAGATCACGG | |||||||
| Tsc21 | 5.08 | 2.92 | 157.67 | 1.00 × 10−10 | Fasciclin domain-containing protein | 617 | F: 5′-TCCAAGACCAGCACCCAATC |
| R: 5′-GGTCTGACAGGGAACTTGGG | |||||||
| Tsc22 | 0.97 | 1.11 | 62.3 | 0.0000006 | Fasciclin domain-containing protein | 525 | F: 5′-CAGGGCTCAACACCCAACAT |
| R: 5′-TAGTTGATGTCGGCAGCCTT | |||||||
| Tsc23 | 1.77 | 1.23 | 12.55 | 2.00 × 10−79 | Cyanophycinase | 1804 | F: 5′-GGAGCTCAGGAGTCGTTGAC |
| R: 5′-CGTTGCGCAAGCTGTAGTAG | |||||||
| Tsc24 | 4.59 | 7.14 | 31.98 | 1.00 × 10−35 | Cathepsin L | 1062 | F: 5′-ACTGACACGGAGGAGATCCA |
| R: 5′-TCCAGTCGACTTTTGTGGGG | |||||||
| Tsc25 | 1.37 | 1.87 | 7.24 | 1.00 × 10−61 | EGF-like domain-containing protein 1 | 2194 | F: 5′-GGAGTGCAACGGAGAGAACA |
| R: 5′-TCTGATGTGTTGGCAGCGAT | |||||||
| Tsc26 | 314.26 | 479.57 | 24.62 | 4.00 × 10−18 | uncharacterized protein; Flags: Fragment | 306 | F: 5′-ATGACTGCCAGGTTTCTCCC |
| R: 5′-CCCAGCAAGCATGTACAAGTG | |||||||
| Tsc27 | 255.69 | 183.9 | 24.21 | 1.00 × 10−16 | Uncharacterized protein | 1000 | F: 5′-ACTGTCTCTACAACAGCGCC |
| R: 5′-GCCTGACTCGAAGATGCAGA | |||||||
| Tsc28 | 188.03 | 95.65 | 16.94 | 3.00 × 10−42 | Uncharacterized protein | 611 | F: 5′-ACAACTGCTCTGCTCCAACA |
| R: 5′-GGTGAGTCTTCCCTTCCCAC | |||||||
| Tsc29 | 156.32 | 133.95 | 8.05 | 3.00 × 10−35 | Uncharacterized protein | 423 | F: 5′-CAGAGCATCTGAGGGAGTGC |
| R: 5′-TGCACTGACCCGAATACACC | |||||||
| Tsc30 | 14.7 | 15.87 | 2.59 | 0 | Dual oxidase | 4918 | F: 5′-CACTTTGATTCCGTCGGGGA |
| R: 5′-CAGCCATGGGTTGATGTCCT | |||||||
| Tsc31 | 10.29 | 12.05 | 2 | 4.00 × 10−61 | Homeobox protein engrailed | 1804 | F: 5′-TGGGTTTTCTGCACTCGCTA |
| R: 5′-TGGAACCGGTGCTCTTCTTC | |||||||
| Tsc32 | 8.17 | 11.22 | 0.83 | 2.00 × 10−167 | Putative ammonium transporter 1 | 1512 | F: 5′-GGACGATTCCACCCGGATAC |
| R: 5′-AAAGGAGATGGCGGCAAAGA | |||||||
| Tsc33 | 72.4 | 52.73 | 6.98 | 2.00 × 10−65 | Annulin | 1013 | F: 5′-GAAACTGGGGCCACGAAGTA |
| R: 5′-CCATGCTTTGACTCCGACCT | |||||||
| Tsc34 | 2.28 | 0.6 | 0 | 0 | Vitelline envelope zona pellucida domain 4 (A0MCN4) | 990 | F: 5′-ATCAGCTGCTACTTCCAGCC |
| R: 5′-ACGGATCCCCATTCACGATG | |||||||
| Tsc35 | 155.23 | 89.83 | 33.46 | 1.00 × 10−32 | Defensin (D3UAH2) | 614 | F: 5′-CTGCTTCTGCTGTGTTTGGT |
| R: 5′-CGACAGACACAGACGCCATT | |||||||
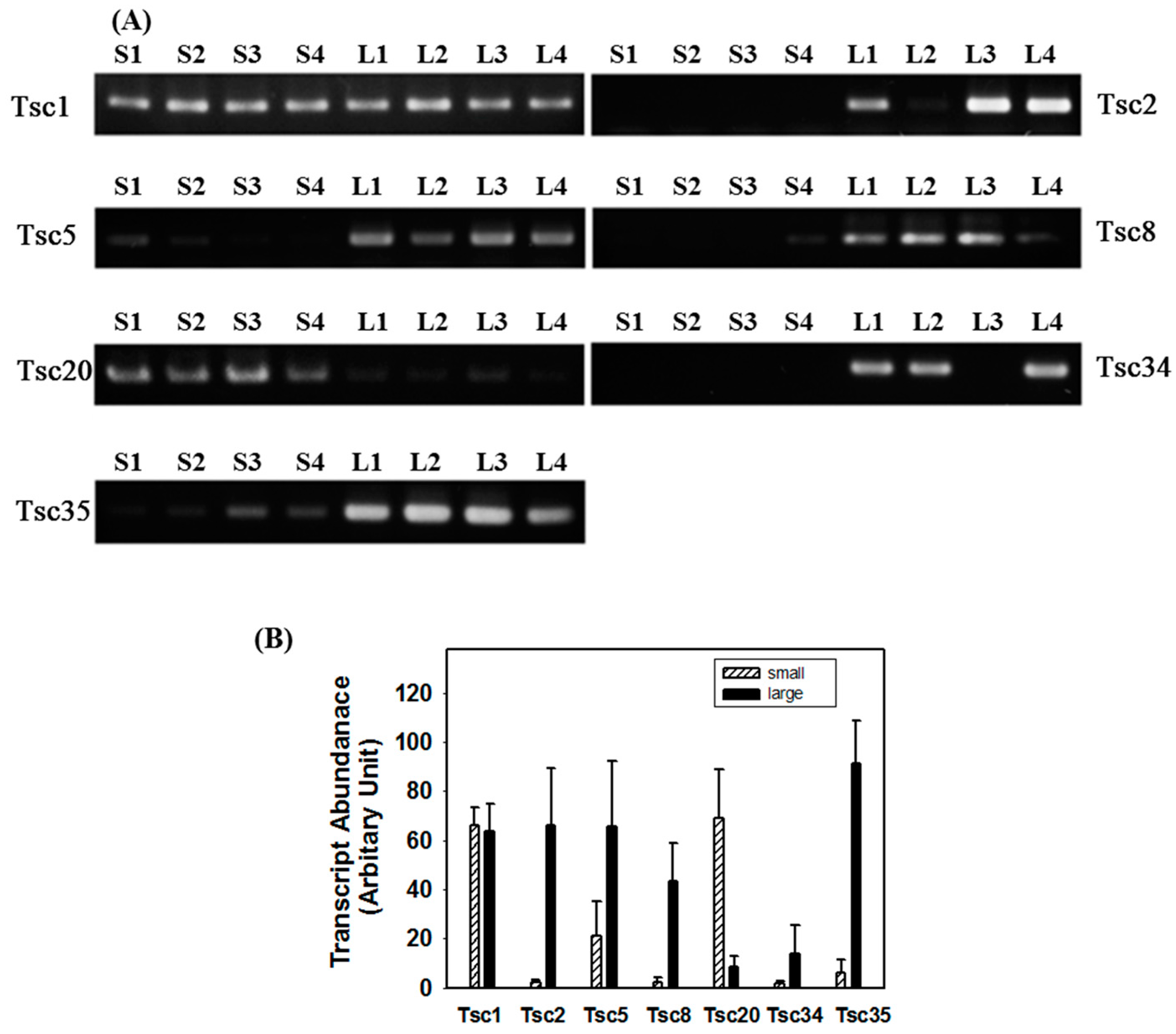
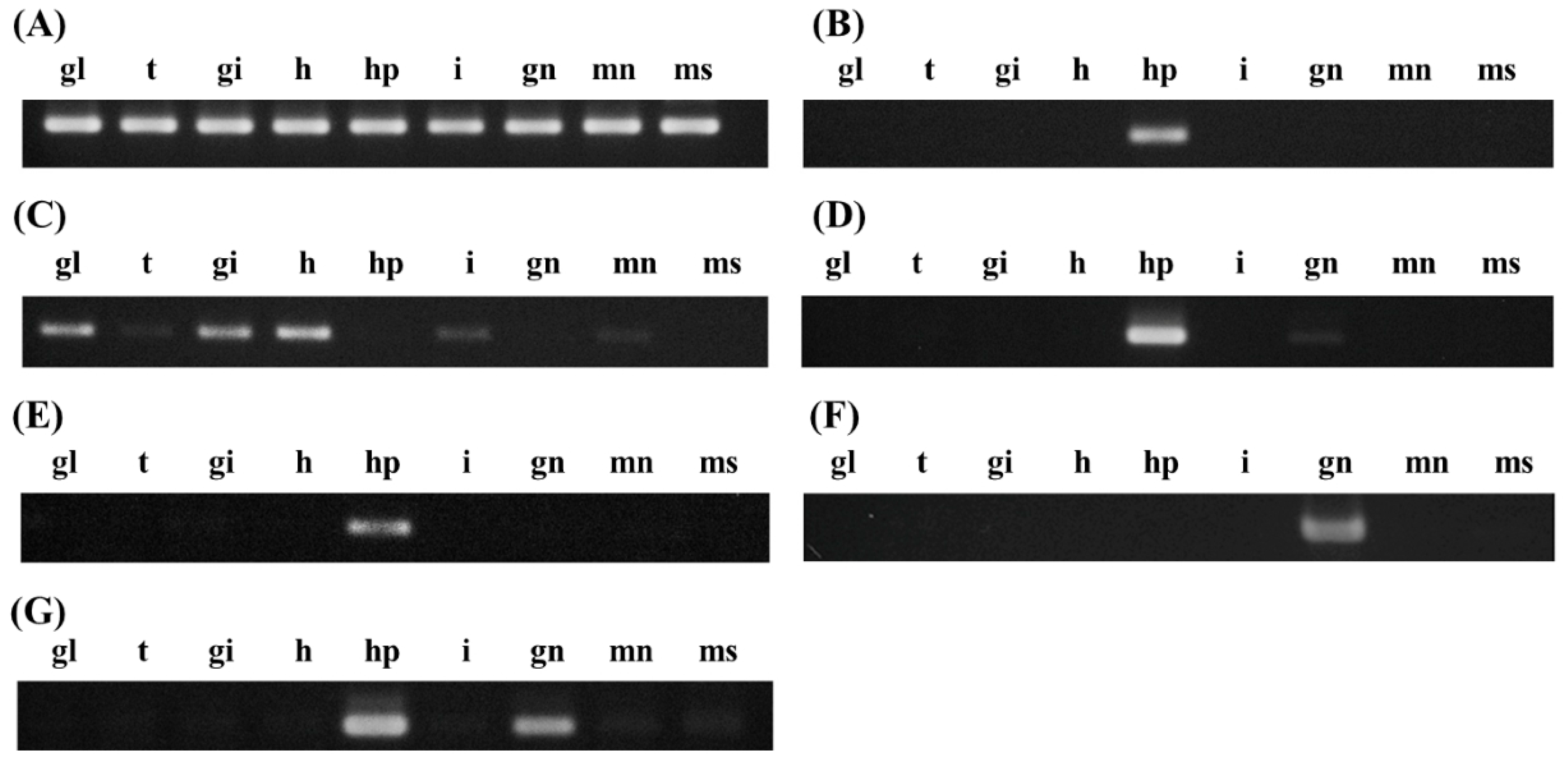
2.4. Tissue-Specific Expression

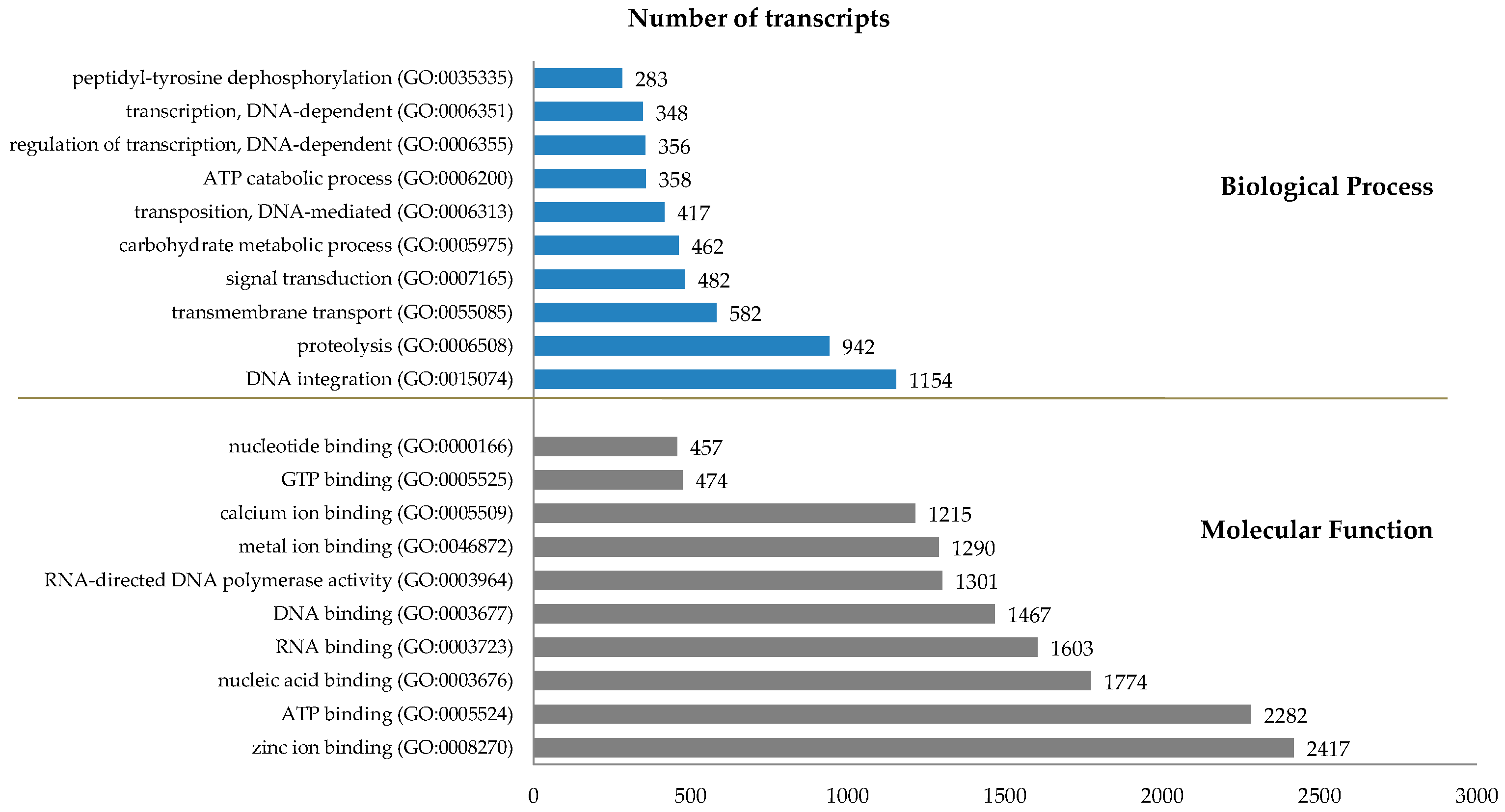
2.5. Functional Implications
3. Experimental Section
3.1. Materials
3.2. RNA Isolation
3.3. Transcriptome Library Construction and Analysis of Differentially-Expressed Genes
3.4. RT-PCR
3.5. Transcript Analysis and Statistics
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gordon, H.R.; Cook, P.A. World abalone fisheries and aquaculture update: Supply and market dynamics. J. Shellfish Res. 2004, 23, 935–939. [Google Scholar]
- Park, C.J.; Kim, S.Y. Abalone aquaculture in Korea. J. Shellfish Res. 2013, 32, 17–19. [Google Scholar] [CrossRef]
- Ock, Y.S. The research on the development procedure and current problems of the Korean abalone industry. J. Fish. Bus. Adm. 2013, 44, 15–28. [Google Scholar] [CrossRef]
- Lopez, L.M.; Tyler, P.; Viana, M.T. The effect of temperature and artificial diets on growth rates of juvenile Haliotis tuberculata (Linnaeus, 1758). J. Shellfish Res. 1998, 17, 657–662. [Google Scholar]
- Nelson, M.M.; Leighton, D.L.; Phleger, C.F.; Nichols, P.D. Comparison of growth and lipid composition in the green abalone, Haliotis fulgens, provided specific macroalgal diets. Comp. Biochem. Physiol. B 2002, 131, 695–712. [Google Scholar] [CrossRef]
- Symonds, J.E.; Moss, G.; Jopson, N.B.; Roberts, R.; Birss, J.; Heath, P.; Walker, S.P.; Anderson, R.M.; McEwan, K.M.; Amer, P.R. Family-based selective breeding of New Zealand abalone, Haliotis iris: Challenges and opportunities. Proc. N. Z. Soc. Anim. 2012, 72, 216–221. [Google Scholar]
- You, W.W.; Ke, C.H.; Luo, X.; Wang, D.X. Growth and survival of three small abalone Haliotis diversicolor populations and their reciprocal crosses. Aquac. Res. 2009, 40, 1474–1480. [Google Scholar] [CrossRef]
- Lucas, T.; Macbeth, M.; Degnan, S.M.; Knibb, W.; Degnan, B.M. Heritability estimates for growth in the tropical abalone Haliotis asinina using microsatellites to assign parentage. Aquaculture 2006, 259, 146–152. [Google Scholar] [CrossRef]
- Baranski, M.; Rourke, M.; Loughnan, S.; Hayes, B.; Austin, C.; Robinson, N. Detection of QTL for growth rate in the blacklip abalone (Haliotis rubra Leach) using selective DNA pooling. Anim. Genet. 2008, 39, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, X.; Zhang, G. Identification of quantitative trait loci for growth-related traits in the Pacific abalone Haliotis discus hannai Ino. Aquac. Res. 2007, 38, 789–797. [Google Scholar] [CrossRef]
- Chen, H.L.; Yang, H.S.; Huang, R.; Tsai, H.-J. Transfer of a foreign gene to Japanese abalone (Haliotis diversicolor supertexta) by direct testis-injection. Aquaculture 2006, 253, 249–258. [Google Scholar] [CrossRef]
- Hamano, K.; Awaji, M.; Usuki, H. cDNA structure of an insulin-related peptide in the Pacific oyster and seasonal changes in the gene expression. J. Endocrinol. 2005, 187, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Geraerts, W.P.M.; Smit, A.B.; Li, K.W.; Hordijk, P.L. The Light Green Cells of Lymnaea: A neuroendocrine model system for stimulus-induced expression of multiple peptide genes in a single cell type. Experientia 1992, 48, 464–473. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; He, M. Differential gene expression identified by RNA-Seq and qPCR in two sizes of pearl oyster (Pinctada fucata). Gene 2014, 538, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Grada, A.; Weinbrecht, K. Next-generation sequencing: Methodology and application. J. Investig. Dermatol. 2013, 133, e11. [Google Scholar] [CrossRef] [PubMed]
- Hegedus, Z.; Zakrzewska, A.; Agoston, V.C.; Ordas, A.; Rácz, P.; Mink, M.; Spaink, H.P.; Meijer, A.H. Deep sequencing of the zebrafish transcriptome response to mycobacterium infection. Mol. Immunol. 2009, 46, 2918–2930. [Google Scholar] [CrossRef] [PubMed]
- Gallardo-Escarate, C.; Valenzuela-Munoz, V.; Nunez-Acuna, G. RNA-Seq analysis using de novo transcriptome assembly as a reference for the Salmon Louse Caligus rogercresseyi. PLoS ONE 2014, 9, e92239. [Google Scholar] [CrossRef] [PubMed]
- Van der Merwe, M.; Franchini, P.; Roodt-Wilding, R. Differential growth-related gene expression in abalone. Mar. Biotechnol. 2011, 13, 1125–1139. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 7, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Ramskold, D.; Wang, E.T.; Burge, C.B.; Sandberg, R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comp. Biol. 2009, 5, e1000598. [Google Scholar] [CrossRef] [PubMed]
- Lira, S.A.; Kinloch, R.A.; Mortillo, S.; Wassarman, P.M. An upstream region of the mouse ZP3 gene directs expression of firefly luciferase specifically to growing oocytes in transgenic mice. Proc. Natl. Acad. Sci. USA 1990, 87, 7215–7219. [Google Scholar] [CrossRef] [PubMed]
- Akatsuka, K.; Yoshida-Komiya, H.; Tulsiani, D.R.P.; Orgebin-Crist, M.-C.; Hiroi, M.; Araki, Y. Rat zona pellucida glycoproteins: Molecular cloning and characterization of the three major components. Mol. Reprod. Dev. 1998, 51, 454–467. [Google Scholar] [CrossRef]
- Drickamer, K. C-type lectin-like domains. Curr. Opin. Struct. Biol. 1999, 9, 585–590. [Google Scholar] [CrossRef]
- Wang, N.; Whang, I.; Lee, J. A novel C-type lectin from abalone, Haliotis discus discus, agglutinates Vibrio alginolyticus. Dev. Comp. Immunol. 2008, 32, 1034–1040. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Qiu, R.; Hu, Y.H. HdhCTL1 is a novel C-type lectin of abalone Haliotis discus hannai that agglutinates Gram-negative bacterial pathogens. Fish Shellfish Immunol. 2014, 41, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, L.; Huang, M.; Zhang, H.; Song, L. The immune role of C-type lectins in molluscs. Invertebr. Surv. J. 2011, 8, 241–246. [Google Scholar]
- Sun, Y.-D.; Fu, L.-D.; Jia, Y.-P.; Du, X.-J.; Wang, Q.; Wang, Y.-H.; Zhao, X.-F.; Yu, X.-Q.; Wang, J.-X. A hepatopancreas-specific C-type lectin from the Chinese shrimp Fenneropenaeus chinensis exhibits antimicrobial activity. Mol. Immunol. 2008, 45, 348–361. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.H.; Benzie, J.A.H.; He, J.-G.; Chan, S.-M. PmLT, a C-type lectin specific to hepatopancreas is involved in the innate defense of the shrimp Penaeus monodon. J. Invertebr. Pathol. 2008, 99, 332–341. [Google Scholar] [PubMed]
- Zhao, Z.-Y.; Yin, Z.-X.; Xu, X.-P.; Weng, S.-P.; Rao, X.-Y.; dai, Z.-X.; Luo, Y.-W.; Yang, G.; Li, Z.-S.; Guan, H.-J.; et al. A novel C-type lectin from the shrimp Litopenaeus vannamei possesses anti-White Spot Syndrome Virus activity. J. Virol. 2009, 83, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-W.; Xu, W.-T.; Wang, X.-W.; Mu, Y.; Zhao, X.-F.; Yu, X.-Q.; Wang, J.-X. A novel C-type lectin with two CRD domains from Chinese shrimp Fenneropenaeus chinensis functions as a pattern recognition protein. Mol. Immunol. 2009, 46, 1626–1637. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-W.; Xu, W.-T.; Zhang, X.-W.; Zhao, X.-F.; Yu, X.-Q.; Wang, J.-X. A C-type lectin is involved in the innate immune response of Chinese white shrimp. Fish Shellfish Immunol. 2009, 27, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.-T.; Wang, X.-W.; Zhang, X.-W.; Zhao, X.-F.; Yu, X.-Q.; Wang, J.-X. A new C-type lectin (FcLec5) from the Chinese white shrimp Fenneropenaeus chinensis. Amino Acids 2010, 39, 1227–1239. [Google Scholar] [CrossRef] [PubMed]
- Weiss, I.M.; Kaufmann, S.; Mann, K.; Fritz, M. Purification and characterization of perlucin and perlustrin, two new proteins from the shell of the mollusc Haliotis laevigata. Biochem. Biophys. Res. Commun. 2000, 267, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Weis, W.I.; Drickamer, K. Structural basis of lectin-carbohydrate recognition. Annu. Rev. Biochem. 1996, 65, 441–473. [Google Scholar] [CrossRef] [PubMed]
- Mann, K.; Weiss, I.M.; André, S.; Gabius, H.-J.; Fritz, M. The amino-acid sequence of the abalone (Haliotis laevigata) nacre protein perlucin. Detection of a functional C-type lectin domain with galactose/mannose specificity. Eur. J. Biochem. 2000, 267, 5257–5264. [Google Scholar] [CrossRef] [PubMed]
- Gardner, L.D.; Mills, D.; Wiegand, A.; Leavesley, D.; Elizur, A. Spatial analysis of biomineralization associated gene expression from the mantle organ of the pearl oyster Pinctada maxima. BMC Genom. 2011, 12, 455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.-Y.; Ma, K.-Y.; Bai, Z.-Y.; Li, J.-L. Molecular cloning and characterization of perlucin from the freshwater pearl mussel, Hyriopsis cumingii. Gene 2013, 526, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Horie, M.; Mitsumoto, Y.; Kyushiki, H.; Kanemoto, N.; Watanabe, A.; Taniguchi, Y.; Nishino, N.; Okamoto, T.; Kondo, M.; Mori, T.; et al. Identification and characterization of TMEFF2, a novel survival factor for hippocampal and mesencephalic neurons. Genomics 2000, 67, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Siegel, D.A.; Huang, M.K.; Becker, S.F. Ectopic dendrite initiation: CNS pathogenesis as a model of CNS development. Int. J. Dev. Neurosci. 2002, 20, 373–389. [Google Scholar] [CrossRef]
- Mohler, J.L.; Morris, T.L.; Ford, O.H., III; Alvey, R.F.; Sakamoto, C.; Gregory, C.W. Identification of differentially expressed genes associated with androgen-independent growth of prostate cancer. Prostate 2002, 51, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Aagaard, J.E.; Yi, X.; MacCoss, M.J.; Swanson, W.J. Rapidly evolving zona pellucida domain proteins are a major component of the vitelline envelope of abalone eggs. Proc. Natl. Acad. Sci. USA 2006, 103, 17302–17307. [Google Scholar] [CrossRef] [PubMed]
- Froy, O.; Gurevitz, M. Arthropod and mollusk defensins—Evolution by exon-shuffling. Trends Genet. 2003, 19, 684–687. [Google Scholar] [CrossRef] [PubMed]
- De Zoysa, M.; Whang, I.; Lee, Y.; Lee, S.; Lee, J.-S.; Lee, J. Defensin from disk abalone Haliotis discus discus: Molecular cloning, sequence characterization and immune response against bacterial infection. Fish Shellfish Immunol. 2010, 28, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Bals, R.; Goldman, M.J.; Wilson, J.M. Mouse beta-defensin 1 is a salt-sensitive antimicrobial peptide present in epithelia of the lung and urogenital tract. Infect. Immun. 1998, 66, 1225–1232. [Google Scholar] [PubMed]
- Selsted, M.E.; Ouellette, A.J. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 2005, 6, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.; Sun, X.; Zheng, M.; Qu, L.; Zan, J.; Zhang, J. Characterization of defensin gene from abalone (Haliotis discus hannai) and its deduced protein. Chin. J. Oceanol. Limnol. 2008, 26, 375–379. [Google Scholar] [CrossRef]
- Park, C.-J.; Lee, J.H.; Noh, J.K.; Kim, H.C.; Park, J.W.; Hwang, I.J.; Kim, S.Y. Growth of pacific abalone, Haliotis discus hannai, using selection breeding technique. Korean J. Malacol. 2012, 28, 343–347. [Google Scholar] [CrossRef]
- Noh, G.E.; Lim, H.K.; Kim, J.-M. Characterization of genes encoding prolactin and prolactin receptors in starry flounder Platichthys stellatus and their expression upon acclimation to freshwater. Fish Physiol. Biochem. 2013, 39, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, M.-J.; Kim, G.-D.; Kim, J.-M.; Lim, H.K. Differentially-Expressed Genes Associated with Faster Growth of the Pacific Abalone, Haliotis discus hannai. Int. J. Mol. Sci. 2015, 16, 27520-27534. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms161126042
Choi M-J, Kim G-D, Kim J-M, Lim HK. Differentially-Expressed Genes Associated with Faster Growth of the Pacific Abalone, Haliotis discus hannai. International Journal of Molecular Sciences. 2015; 16(11):27520-27534. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms161126042
Chicago/Turabian StyleChoi, Mi-Jin, Gun-Do Kim, Jong-Myoung Kim, and Han Kyu Lim. 2015. "Differentially-Expressed Genes Associated with Faster Growth of the Pacific Abalone, Haliotis discus hannai" International Journal of Molecular Sciences 16, no. 11: 27520-27534. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms161126042






