Metabolite Profile of Cervicovaginal Fluids from Early Pregnancy Is Not Predictive of Spontaneous Preterm Birth
Abstract
:1. Introduction
Study Design
| Characteristic | sPTB Cases (n = 30) | Controls (n = 30) |
|---|---|---|
| Maternal Age (years) a | 32 ± 3.2 | 32 ± 3.5 |
| Maternal BMI (Body Mass Index) a | 24.9 ± 4.5 | 24.7 ± 3.5 |
| Ethnicity | 27 Caucasians, 3 Asians | 27 Caucasians, 3 Asians |
| Gestational age at the delivery (weeks) | 34.3 ± 2.0 * | 39.9 ± 0.9 * |
| Gestational age at sampling (weeks) | 20.1 ± 0.7 | 19.6 ± 0.8 |
| Cigarette smokers a | 29/30 did not smoke at recruitment | None smoked at recruitment |
| Shortest transvaginal cervical length at 20 weeks gestation (mm) | 40.3 ± 8.9 | 39.5 ± 5.7 |
| PPROM | 11/30 | - |
| Previous termination at >10 weeks’ | 1/30 | - |
| Vaginal bleeding during pregnancy b | 8/30 | 4/30 |
| Proven vaginal candida infection in pregnancy b | 2/30 | 2/30 |
| Any infection during pregnancy (gasteroentiritis, pyelonephritis, vaginal candida b | 17/30 | 12/30 |
| Fertility treatment to conceive current pregnancy | 7/30 | 1/30 |
2. Results and Discussion
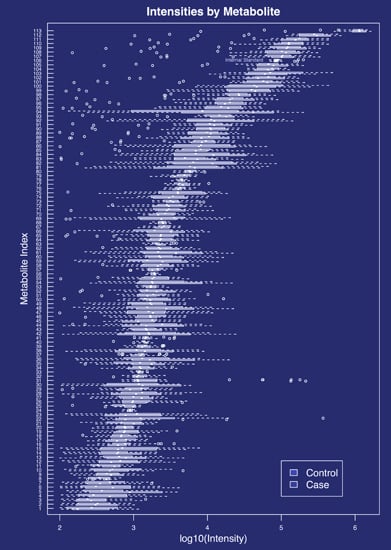
2.1. Metabolites Present in the Cervicovaginal Fluid
2.2. Inflammatory Markers, Infection and Metabolite Profile
| Marker | Infection (p) | Infection (q *) | sPTB-control (p) | sPTB-control (q) |
|---|---|---|---|---|
| CXCL10 16a | 0.002 | 0.029 | 0.889 | 0.889 |
| IL-1ra31a | 0.008 | 0.046 | 0.181 | 0.363 |
| TNFR1A 80b | 0.079 | 0.236 | 0.669 | 0.803 |
| MMP9 51a | 0.151 | 0.362 | 0.530 | 0.794 |
| TIMP-1 126 | 0.278 | 0.477 | 0.782 | 0.853 |
| CRP 120 | 0.644 | 0.803 | 0.057 | 0.229 |
2.3. Discussion
3. Experimental Section
3.1. Sample Collection
3.2. Sample Selection and Randomisation
3.3. GC-MS Sample Preparation
3.4. GC-MS Data Acquisition
3.5. GC-MS Data Extraction and Compound Identification
3.6. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Blencowe, H.; Cousens, S.; Chou, D.; Oestergaard, M.; Say, L.; Moller, A.-B.; Kinney, M.; Lawn, J. Born too soon: The global epidemiology of 15 million preterm births. Reprod. Health 2013, 10 (Suppl. 1), S2. [Google Scholar] [CrossRef] [PubMed]
- Blencowe, H.; Cousens, S.; Oestergaard, M.Z.; Chou, D.; Moller, A.-B.; Narwal, R.; Lawn, J.E. National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: A systematic analysis and implications. Lancet 2012, 379, 2162–2172. [Google Scholar] [CrossRef]
- Ball, G.; Boardman, J.P.; Aljabar, P.; Pandit, A.; Arichi, T.; Merchant, N.; Rueckert, D.; Edwards, A.D.; Counsell, S.J. The influence of preterm birth on the developing thalamocortical connectome. Cortex 2013, 49, 1711–1721. [Google Scholar] [CrossRef] [PubMed]
- Nosarti, C.; Nam, K.W.; Walshe, M.; Murray, R.M.; Cuddy, M.; Rifkin, L.; Allin, M.P.G. Preterm birth and structural brain alterations in early adulthood. Neuroimage Clin. 2014, 6, 180–191. [Google Scholar] [CrossRef] [PubMed]
- Crump, C.; Winkleby, M.A.; Sundquist, K.; Sundquist, J. Risk of diabetes among young adults born preterm in Sweden. Diabetes Care 2011, 34, 1109–1113. [Google Scholar] [CrossRef] [PubMed]
- Auger, N.; Abrahamowicz, M.; Wynant, W.; Lo, E. Gestational age-dependent risk factors for preterm birth: Associations with maternal education and age early in gestation. Eur. J. Obetet. Gynecol. Reprod. Biol. 2014, 176, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Iams, J.D. Clinical Practice: Prevention of preterm parturition. N. Engl. J. Med. 2014, 370, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Menon, R.; Jones, J.; Gunst, P.R.; Kacerovsky, M.; Fortunato, S.J.; Saade, G.R.; Basraon, S. Amniotic fluid metabolomic analysis in spontaneous preterm birth. Reprod. Sci. 2014, 21, 791–803. [Google Scholar] [CrossRef] [PubMed]
- Conde-Agudelo, A.; Papageorghiou, A.T.; Kennedy, S.H.; Villar, J. Novel biomarkers for the prediction of the spontaneous preterm birth phenotype: A systematic review and meta-analysis. Eur. J. Obetet. Gynecol. Reprod. Biol. 2011, 118, 1042–1054. [Google Scholar] [CrossRef] [PubMed]
- Honest, H.; Bachmann, L.M.; Gupta, J.K.; Kleijnen, J.; Khan, K.S. Accuracy of cervicovaginal fetal fibronectin test in predicting risk of spontaneous preterm birth: Systematic review. BMJ 2002, 325, 301–304. [Google Scholar] [CrossRef] [PubMed]
- Stout, M.J.; Goetzinger, K.R.; Tuuli, M.G.; Cahill, A.G.; Macones, G.A.; Odibo, A.O. First trimester serum analytes, maternal characteristics and ultrasound markers to predict pregnancies at risk for preterm birth. Placenta 2012, 34, 14–19. [Google Scholar] [CrossRef] [PubMed]
- McDonald, C.R.; Darling, A.M.; Conroy, A.L.; Tran, V.; Cabrera, A.; Liles, W.C.; Wang, M.; Aboud, S.; Urassa, W.; Fawzi, W.W.; et al. Inflammatory and angiogenic factors at mid-pregnancy are associated with spontaneous preterm birth in a cohort of Tanzanian Women. PLoS ONE 2015. [Google Scholar] [CrossRef]
- Maitre, L.; Fthenou, E.; Athersuch, T.; Coen, M.; Toledano, M.; Holmes, E.; Kogevinas, M.; Chatzi, L.; Keun, H. Urinary metabolic profiles in early pregnancy are associated with preterm birth and fetal growth restriction in the Rhea mother-child cohort study. BMC Med. 2014, 12, 110. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Tanada, C.; Sakamoto, T.; Tsutsui, H.; Akiba, T.; Min, J.Z.; Todoroki, K.; Yamano, Y.; Toyo Oka, T. Metabolomics approach of infant formula for the evaluation of contamination and degradation using hydrophilic interaction liquid chromatography coupled with mass spectrometry. Food Chem. 2015, 181, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Power, K.M.; Sanchez-Galan, J.E.; Luskey, G.W.; Koski, K. G.; Burns, D.H. Use of near-infrared spectroscopic analysis of second trimester amniotic fluid to assess preterm births. J. Pregnancy 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Liong, S.; di Quinzio, M.K.W.; Fleming, G.; Permezel, M.; Rice, G.E.; Georgiou, H.M. Prediction of spontaneous preterm labour in at-risk pregnant women. Reproduction 2013, 146, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Alleman, B.W.; Smith, A.R.; Byers, H.M.; Bedell, B.; Ryckman, K.K.; Murray, J.C.; Borowski, K.S. A proposed method to predict preterm birth using clinical data, standard maternal serum screening, and cholesterol. Am. J. Obstet. Gynecol. 2013, 208, e471–e472. [Google Scholar] [CrossRef] [PubMed]
- Heazell, A.E.P.; Bernatavicius, G.; Warrander, L.; Brown, M.C.; Dunn, W.B. A metabolomic approach identifies differences in maternal serum in third trimester pregnancies that end in poor perinatal outcome. Reprod. Sci. 2012, 19, 863–875. [Google Scholar] [CrossRef] [PubMed]
- Utts, J.M.; Heckard, R.F. Statistical Ideas and Methods; Thompson Brooks/Cole: Belmont, CA, USA, 2006. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, Y.; Romero, R.; Mele, L.; Wapner, R.J.; Iams, J.D.; Dudley, D.J.; Spong, C.Y.; Peaceman, A.M.; Leveno, K.J.; Harper, M.; et al. Maternal serum interleukin-6, C-reactive protein, and matrix metalloproteinase-9 concentrations as risk factors for preterm birth <32 weeks and adverse neonatal outcomes. Am. J. Perinatol. 2010, 27, 631–640. [Google Scholar] [PubMed]
- Combs, C.A.; Gravett, M.; Garite, T.J.; Hickok, D.E.; Lapidus, J.; Porreco, R.; Rael, J.; Grove, T.; Morgan, T.K.; Clewell, W.; et al. Amniotic fluid infection, inflammation, and colonization in preterm labor with intact membranes. Am. J. Obstet. Gynecol. 2014, 210, e1–e125. [Google Scholar] [CrossRef] [PubMed]
- Hollander, M.; Wolfe, D.A. Nonparametric Statistical Methods; John Wiley & Sons: New York, NY, USA, 1973; pp. 115–120. [Google Scholar]
- Villas-Bôas, S.G.; Smart, K.F.; Sivakumaran, S.; Lane, G.A. Alkylation or silylation for analysis of amino and non-amino organic acids by GC-MS? Metabolites 2011, 1, 3–20. [Google Scholar] [CrossRef] [PubMed]
- De Seymour, J.V.; Conlon, C.A.; Sulek, K.; Villas Bôas, S.G.; McCowan, L.M.E.; Kenny, L.C.; Baker, P.N. Early pregnancy metabolite profiling discovers a potential biomarker for the subsequent development of gestational diabetes mellitus. Acta Diabetol. 2014, 51, 887–890. [Google Scholar] [CrossRef] [PubMed]
- Auray-Blais, C.; Raiche, E.; Gagnon, R.; Berthiaume, M.; Pasquier, J.-C. Metabolomics and preterm birth: What biomarkers in cervicovaginal secretions are predictive of high-risk pregnant women? Int. J. Mol. Sci. 2011, 307, 33–38. [Google Scholar] [CrossRef]
- Kell, D.B. Metabolomic biomarkers: search, discovery and validation. Expert Rev. Mol. Diagn. 2007, 7, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Isaaq, H.L.; Veenstra, T.D. Analytical methods and biomarker validation. In Proteomic and Metabolomic Approaches to Biomarker Discovery; Elsevier: London, UK, 2013; pp. 447–450. [Google Scholar]
- Broadhurst, D.I.; Kell, B.D. Statistical strategies for avoiding false discoveries in metabolomics and related experiments. Metabolomics 2006, 2, 171–196. [Google Scholar] [CrossRef]
- Ghartey, J.; Bastek, J.A.; Brown, A.G.; Anglim, L.; Elovitz, M.A. Women with preterm birth have a distinct cervicovaginal metabolome. Am. J. Obstet. Gynecol. 2015, 212, 776e1–776e12. [Google Scholar] [CrossRef] [PubMed]
- Ayache, S.; Panelli, M.; Marincola, M.F.; Stroncek, D.F. Effects of storage time and exogenous protease inhibitors on plasma protein levels. Am. J. Clin. Pathol. 2006, 126, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Smart, K.F.; Aggio, R.B.M.; van Houtte, J.R.; Villas-Bôas, S.G. Analytical platform for metabolome analysis of microbial cells using methyl chloroformate derivatization followed by gas chromatography-mass spectrometry. Nat. Protoc. 2010, 5, 1709–1729. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thomas, M.M.; Sulek, K.; McKenzie, E.J.; Jones, B.; Han, T.-L.; Villas-Boas, S.G.; Kenny, L.C.; McCowan, L.M.E.; Baker, P.N. Metabolite Profile of Cervicovaginal Fluids from Early Pregnancy Is Not Predictive of Spontaneous Preterm Birth. Int. J. Mol. Sci. 2015, 16, 27741-27748. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms161126052
Thomas MM, Sulek K, McKenzie EJ, Jones B, Han T-L, Villas-Boas SG, Kenny LC, McCowan LME, Baker PN. Metabolite Profile of Cervicovaginal Fluids from Early Pregnancy Is Not Predictive of Spontaneous Preterm Birth. International Journal of Molecular Sciences. 2015; 16(11):27741-27748. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms161126052
Chicago/Turabian StyleThomas, Melinda M., Karolina Sulek, Elizabeth J. McKenzie, Beatrix Jones, Ting-Li Han, Silas G. Villas-Boas, Louise C. Kenny, Lesley M. E. McCowan, and Philip N. Baker. 2015. "Metabolite Profile of Cervicovaginal Fluids from Early Pregnancy Is Not Predictive of Spontaneous Preterm Birth" International Journal of Molecular Sciences 16, no. 11: 27741-27748. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms161126052