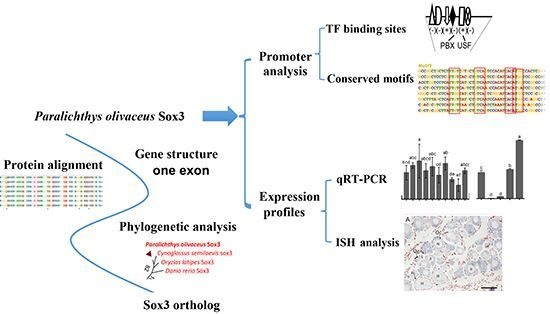
Molecular Cloning, Promoter Analysis and Expression Profiles of the sox3 Gene in Japanese Flounder, Paralichthys olivaceus
Abstract
:1. Introduction
2. Results
2.1. Molecular Characterization of Posox3
2.1.1. Cloning and Sequence Analysis of Posox3

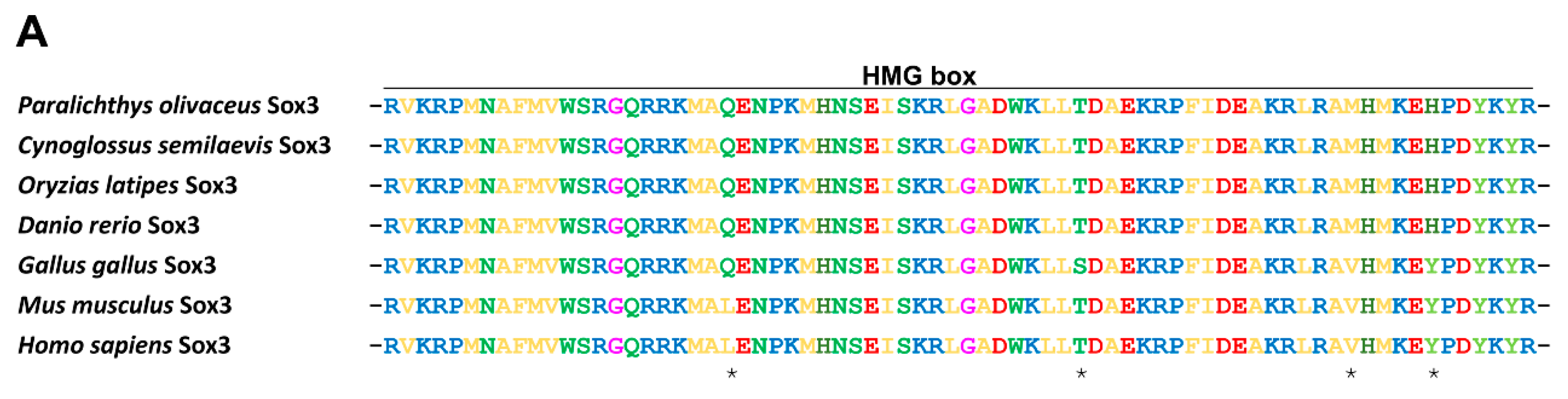
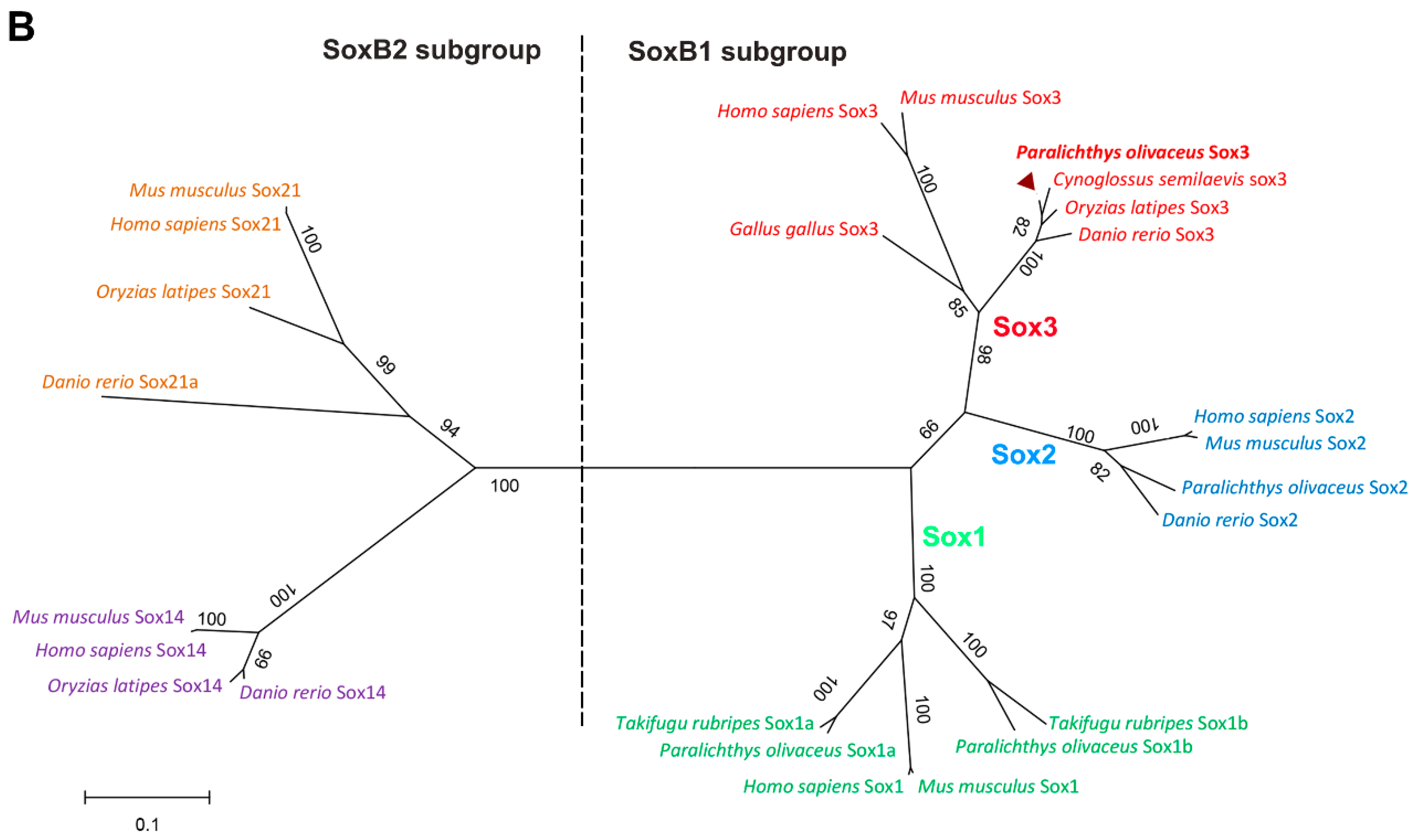
2.1.2. Protein Alignment and Phylogenetic Analysis of PoSox3
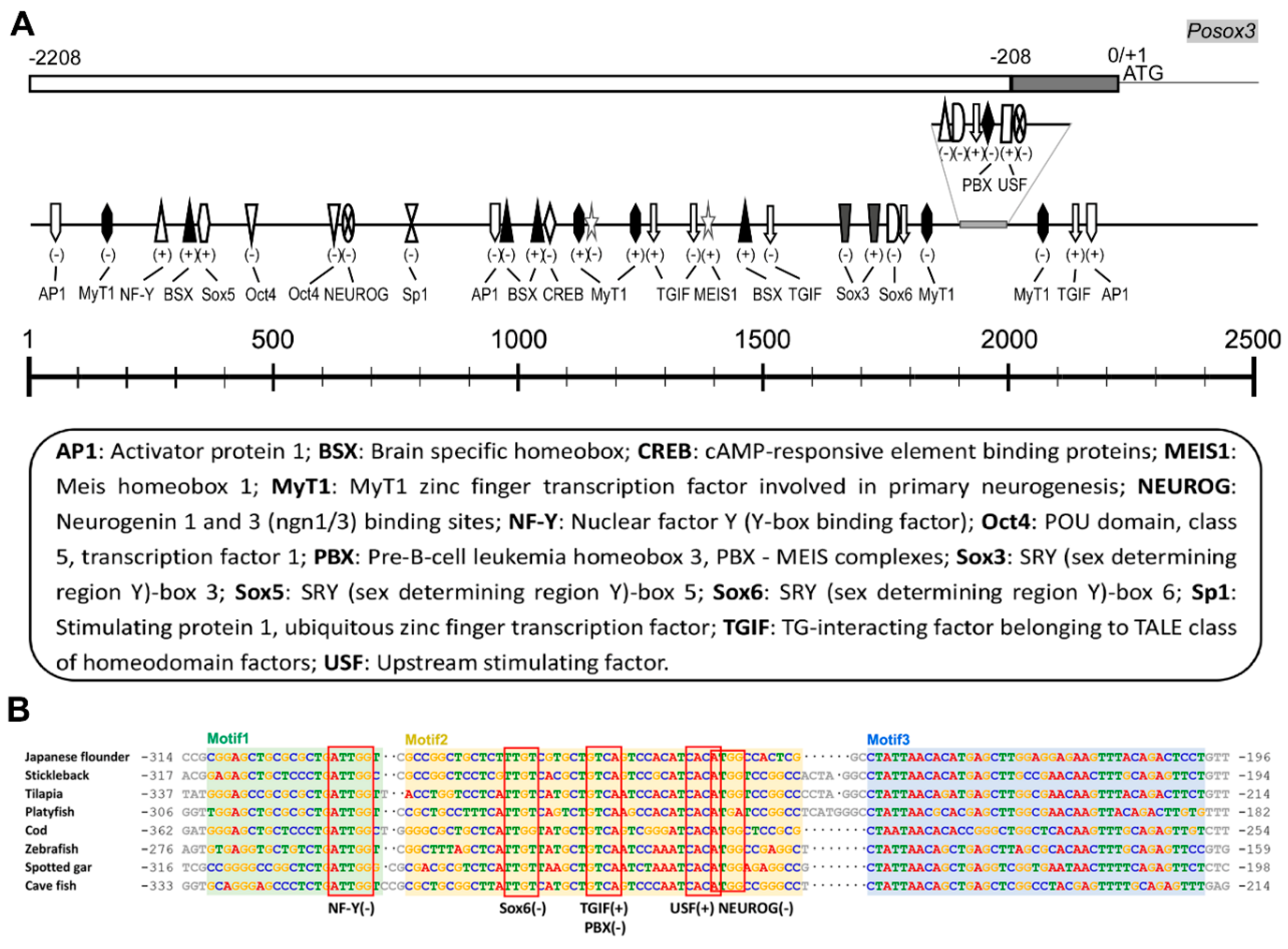
2.1.3. Genomic Organization and Analysis of Posox3
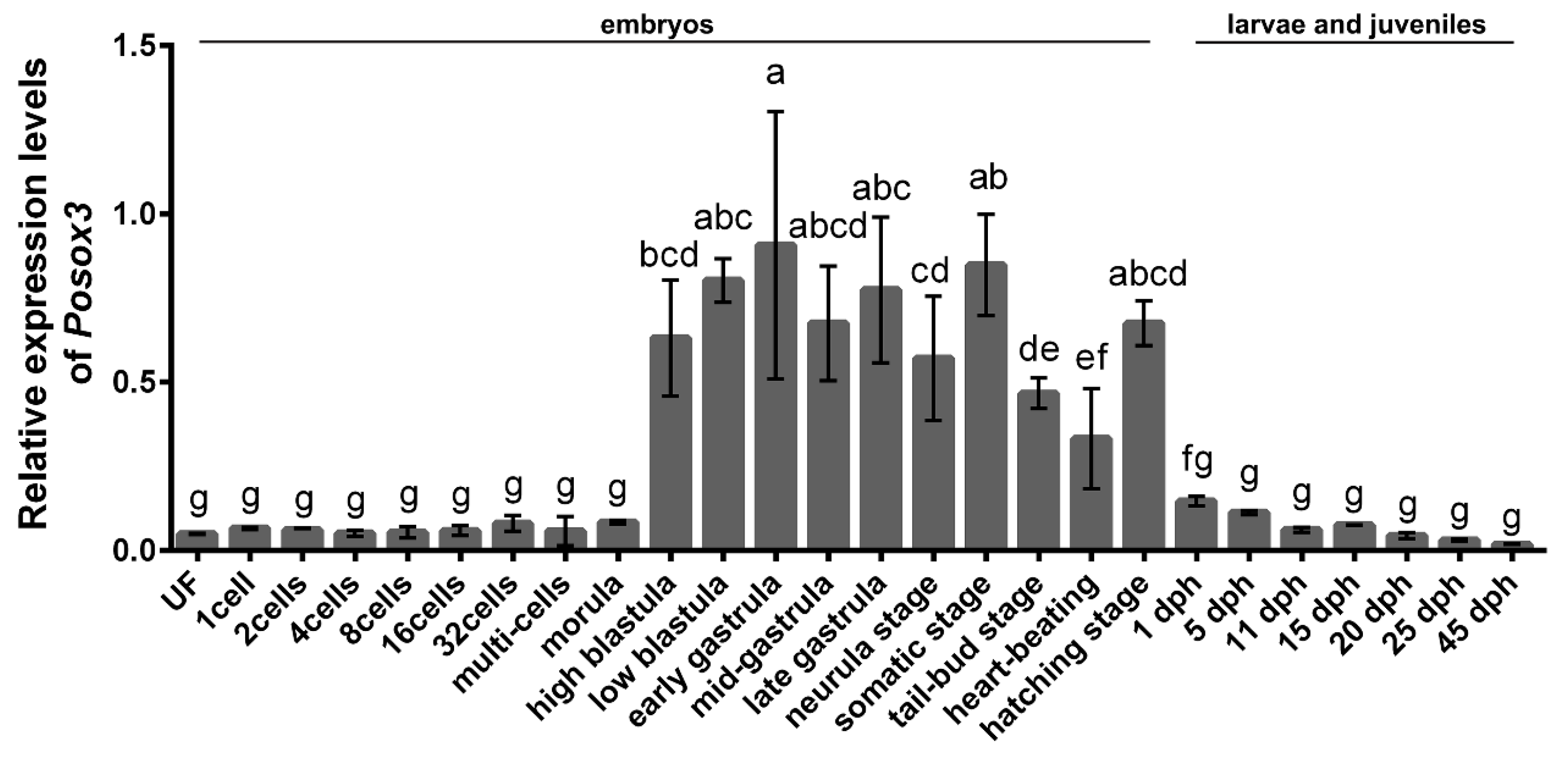
2.2. Expression Profiles of Posox3 during Embryonic Development and in the Larval and Juveniles
2.3. Expression Profiles of Posox3 in Different Tissues and in Adult Brain

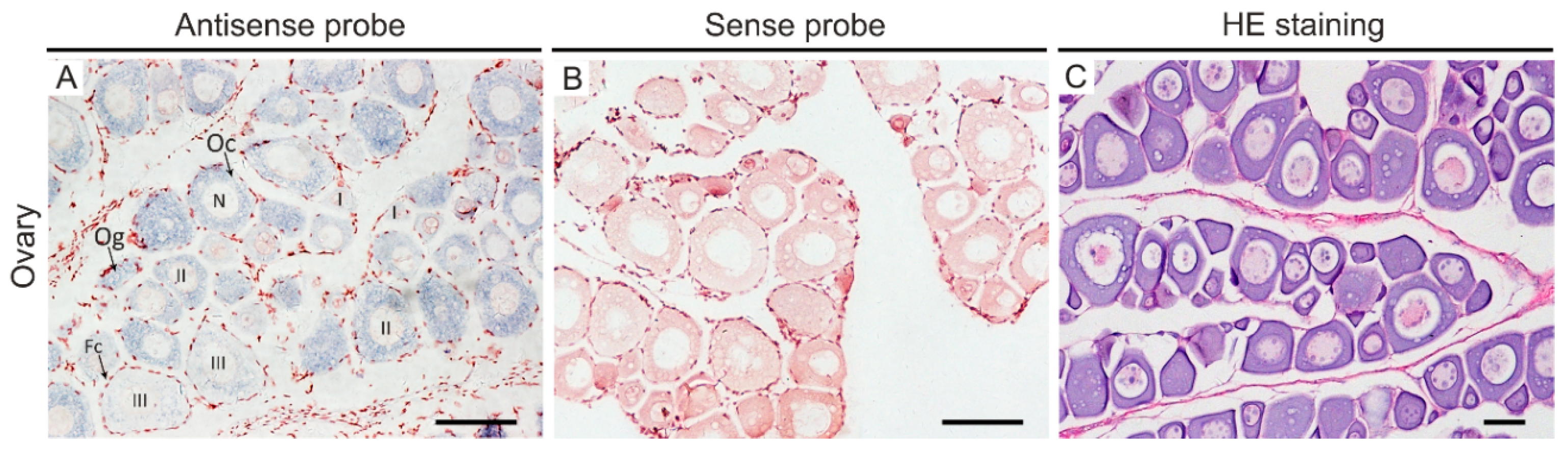
2.4. In Situ Hybridization
3. Discussion
3.1. Posox3 Is the Homolog of Mammalian Sox3
3.2. The Potential TFs in Regulating Posox3 Expression
3.3. Posox3 May Be Involved in the Regulation of Embryogenesis, Neural and Ovarian Development
4. Experimental Section
4.1. Animals and Sampling
4.2. Total RNA and Genomic DNA Extraction
| Experiment | Primer Name | Primer Sequences (5′–3′) |
|---|---|---|
| DNA detection | β-actin-Fw | GAGATGAAGCCCAGAGCAAGAG |
| β-actin-Rv | CAGCTGTGGTGGTGAAGGAGTAG | |
| Core fragment | sox3-core-Fw | AGCCGCCTCAATGTATAGCA |
| sox3-core-Rv | GCGTAACCGTCCATCCTCT | |
| 5′ RACE PCR | 5′ RACE-sox3-467 | GTCGGTCAGAAGTTTCCAGTCAG |
| 5′ RACE-sox3-327 | GGTCGCTGGCACTGTTGTTCT | |
| 3′ RACE PCR | 3′ RACE-sox3-654 | CAGAGGATGGACGGTTACGC |
| 3′ RACE-sox3-827 | TACACGCAGCAGACCACCAAC | |
| Full-length cDNA | sox3-full-length-Fw | AACATTGAACATGATTACGATTCG |
| sox3-full-length-Rv | ATCGTACAAATTTATTCAGCATTAG | |
| Promoter | sox3-promoter-Fw | GCCTGTATTTGTAGTCTAAT |
| sox3-promoter-Rv | TGAGGCGGCTTTAAGTGTT | |
| qRT-PCR | sox3-RT-Fw | AACAACGCAGTGTCGGTGG |
| sox3-RT-Rv | TGCTGTGATGCTGAGGGTAGG | |
| qRT-PCR | 18S-RT-Fw | GGTAACGGGGAATCAGGGT |
| 18S-RT-Rv | TGCCTTCCTTGGATGTGGT | |
| qRT-PCR | UbcE-RT-Fw | TTACTGTCCATTTCCCCACTGAC |
| UbcE-RT-Rv | GACCACTGCGACCTCAAGATG | |
| ISH | sox3-ISH-SP6 | ATTTAGGTGACACTATAGAAGTG |
| TACACGCAGCAGACCACCAAC | ||
| ISH | sox3-ISH-T7 | TAATACGACTCACTATAGGGAGA |
| AAATACACCAGCCTCCTCACG |
4.3. Molecular Cloning of the Full Length cDNA and Promoter Sequence of Posox3
4.4. Sequence Analysis
4.5. Quantitative Real-Time RT-PCR (qRT-PCR)
4.6. In Situ Hybridization and Histological Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bowles, J.; Schepers, G.; Koopman, P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 2000, 227, 239–255. [Google Scholar] [CrossRef] [PubMed]
- Rogers, C.D.; Harafuji, N.; Archer, T.; Cunningham, D.D.; Casey, E.S. Xenopus Sox3 activates Sox2 and geminin and indirectly represses Xvent2 expression to induce neural progenitor formation at the expense of non-neural ectodermal derivatives. Mech. Dev. 2009, 126, 42–55. [Google Scholar] [CrossRef] [PubMed]
- Okuda, Y.; Ogura, E.; Kondoh, H.; Kamachi, Y. B1 SOX coordinate cell specification with patterning and morphogenesis in the early zebrafish embryo. PLoS Genet. 2010, 6, e1000936. [Google Scholar] [CrossRef] [PubMed]
- Wood, H.B.; Episkopou, V. Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech. Dev. 1999, 86, 197–201. [Google Scholar] [CrossRef]
- Elkouris, M.; Balaskas, N.; Poulou, M.; Politis, P.K.; Panayiotou, E.; Malas, S.; Thomaidou, D.; Remboutsika, E. Sox1 maintains the undifferentiated state of cortical neural progenitor cells via the suppression of prox1-mediated cell cycle exit and neurogenesis. Stem Cells 2011, 29, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Graham, V.; Khudyakov, J.; Ellis, P.; Pevny, L. Sox2 functions to maintain neural progenitor identity. Neuron 2003, 39, 749–765. [Google Scholar] [CrossRef]
- Wang, T.W.; Stromberg, G.P.; Whitney, J.T.; Brower, N.W.; Klymkowsky, M.W.; Parent, J.M. Sox3 expression identifies neural progenitors in persistent neonatal and adult mouse forebrain germinative zones. J. Comp. Neurol. 2006, 497, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Bylund, M.; Andersson, E.; Novitch, B.G.; Muhr, J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat. Neurosci. 2003, 6, 1162–1168. [Google Scholar] [CrossRef] [PubMed]
- Avilion, A.A.; Nicolis, S.K.; Pevny, L.H.; Perez, L.; Vivian, N.; Lovell-Badge, R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003, 17, 126–140. [Google Scholar] [CrossRef] [PubMed]
- Adachi, K.; Suemori, H.; Yasuda, S.Y.; Nakatsuji, N.; Kawase, E. Role of sox2 in maintaining pluripotency of human embryonic stem cells. Genes Cells 2010, 15, 455–470. [Google Scholar] [PubMed]
- Kan, L.; Israsena, N.; Zhang, Z.; Hu, M.; Zhao, L.R.; Jalali, A.; Sahni, V.; Kessler, J.A. Sox1 acts through multiple independent pathways to promote neurogenesis. Dev. Biol. 2004, 269, 580–594. [Google Scholar] [CrossRef] [PubMed]
- Weiss, J.; Meeks, J.J.; Hurley, L.; Raverot, G.; Frassetto, A.; Jameson, J.L. Sox3 is required for gonadal function, but not sex determination, in males and females. Mol. Cell. Biol. 2003, 23, 8084–8091. [Google Scholar] [CrossRef] [PubMed]
- Raverot, G.; Weiss, J.; Park, S.Y.; Hurley, L.; Jameson, J.L. Sox3 expression in undifferentiated spermatogonia is required for the progression of spermatogenesis. Dev. Biol. 2005, 283, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Okuda, Y.; Yoda, H.; Uchikawa, M.; Furutani-Seiki, M.; Takeda, H.; Kondoh, H.; Kamachi, Y. Comparative genomic and expression analysis of group B1 sox genes in zebrafish indicates their diversification during vertebrate evolution. Dev. Dyn. 2006, 235, 811–825. [Google Scholar] [CrossRef] [PubMed]
- Dee, C.T.; Hirst, C.S.; Shih, Y.H.; Tripathi, V.B.; Patient, R.K.; Scotting, P.J. Sox3 regulates both neural fate and differentiation in the zebrafish ectoderm. Dev. Biol. 2008, 320, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Yao, B.; Zhou, L.; Wang, Y.; Xia, W.; Gui, J.F. Differential expression and dynamic changes of SOX3 during gametogenesis and sex reversal in protogynous hermaphroditic fish. J. Exp. Zool. A 2007, 307, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Rajakumar, A.; Senthilkumaran, B. Expression analysis of sox3 during testicular development, recrudescence, and after hCG induction in catfish, clarias batrachus. Sex. Dev. 2014, 8, 376–386. [Google Scholar] [PubMed]
- Takehana, Y.; Matsuda, M.; Myosho, T.; Suster, M.L.; Kawakami, K.; Shin, I.T.; Kohara, Y.; Kuroki, Y.; Toyoda, A.; Fujiyama, A.; et al. Co-option of Sox3 as the male-determining factor on the y chromosome in the fish Oryzias dancena. Nat. Commun. 2014, 5, 4157. [Google Scholar] [CrossRef] [PubMed]
- Kovacevic Grujicic, N.; Mojsin, M.; Krstic, A.; Stevanovic, M. Functional characterization of the human SOX3 promoter: Identification of transcription factors implicated in basal promoter activity. Gene 2005, 344, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Krstic, A.; Mojsin, M.; Stevanovic, M. Regulation of SOX3 gene expression is driven by multiple NF-y binding elements. Arch. Biochem. Biophys. 2007, 467, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Mojsin, M.; Popovic, J.; Kovacevic Grujicic, N.; Stevanovic, M. Tg-interacting factor (TGIF) downregulates SOX3 gene expression in the NT2/D1 cell line. J. Genet. Genom. 2012, 39, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Kovacevic-Grujicic, N.; Mojsin, M.; Popovic, J.; Petrovic, I.; Topalovic, V.; Stevanovic, M. Cyclic AMP response element binding (CREB) protein acts as a positive regulator of SOX3 gene expression in NT2/D1 cells. BMB Rep. 2014, 47, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Mojsin, M.; Stevanovic, M. PBX1 and MEIS1 up-regulate SOX3 gene expression by direct interaction with a consensus binding site within the basal promoter region. Biochem. J. 2010, 425, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Savare, J.; Bonneaud, N.; Girard, F. SUMO represses transcriptional activity of the drosophila soxneuro and human sox3 central nervous system-specific transcription factors. Mol. Biol. Cell 2005, 16, 2660–2669. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, Z.; Shao, K.; Fan, L.; Yang, L.; Song, H.; Liu, M.; Wang, Z.; Wang, X.; Zhang, Q. Identification and characterization of a Sox2 homolog in the japanese flounder paralichthys olivaceus. Gene 2014, 544, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Zhang, W.; Li, P.; Liu, J.; Song, H.; Wang, X.; Zhang, Q. Identification, molecular characterization and gene expression analysis of sox1a and sox1b gene in japanese flounder, paralichthys olivaceus. Gene 2015, 574, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Kovacevic-Grujicic, N.; Mojsin, M.; Djurovic, J.; Petrovic, I.; Stevanovic, M. Comparison of promoter regions of SOX3, SOX14 and SOX18 orthologs in mammals. DNA Seq. 2008, 19, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Ensembl. Available online: http://asia.ensembl.org/index.html. (assessed on 26 August 2015).
- Stevanovic, M.; Lovell-Badge, R.; Collignon, J.; Goodfellow, P.N. Sox3 is an x-linked gene related to sry. Hum. Mol. Genet. 1993, 2, 2013–2018. [Google Scholar] [CrossRef] [PubMed]
- Koyano, S.; Ito, M.; Takamatsu, N.; Takiguchi, S.; Shiba, T. The xenopussox3 gene expressed in oocytes of early stages. Gene 1997, 188, 101–107. [Google Scholar] [CrossRef]
- Wiebe, M.S.; Nowling, T.K.; Rizzino, A. Identification of novel domains within Sox-2 and Sox-11 involved in autoinhibition of DNA binding and partnership specificity. J. Biol. Chem. 2003, 278, 17901–17911. [Google Scholar] [CrossRef] [PubMed]
- Aota, S.; Nakajima, N.; Sakamoto, R.; Watanabe, S.; Ibaraki, N.; Okazaki, K. Pax6 autoregulation mediated by direct interaction of Pax6 protein with the head surface ectoderm-specific enhancer of the mouse Pax6 gene. Dev. Biol. 2003, 257, 1–13. [Google Scholar] [CrossRef]
- Frankenberg, S.; Pask, A.; Renfree, M.B. The evolution of class V POU domain transcription factors in vertebrates and their characterisation in a marsupial. Dev. Biol. 2010, 337, 162–170. [Google Scholar] [CrossRef] [PubMed]
- McAninch, D.; Thomas, P. Identification of highly conserved putative developmental enhancers bound by SOX3 in neural progenitors using CHIP-Seq. PLoS ONE 2014, 9, e113361. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, J.; Jiang, J.; Fan, L.; Wang, W.; Liu, J.; Zhang, Q.; Wang, X. Identification and characterization of a nanog homolog in japanese flounder (paralichthys olivaceus). Gene 2013, 531, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Weirauch, M.T.; Hughes, T.R. Conserved expression without conserved regulatory sequence: The more things change, the more they stay the same. Trends Genet. 2010, 26, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Archer, T.C.; Jin, J.; Casey, E.S. Interaction of Sox1, Sox2, Sox3 and Oct4 during primary neurogenesis. Dev. Biol. 2011, 350, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Djurovic, J.; Stevanovic, M. Structural and functional characterization of the human SOX14 promoter. Biochim. Biophys. Acta 2004, 1680, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Bo, Y.; Li, Z.; Jianfang, G. Studies on cDNA cloning and temporal and spatial expression of sox3 gene in grouper epinephelus coioides. High Technol. Lett. 2003, 13, 74–81. [Google Scholar]
- Wagner, J.T.; Podrabsky, J.E. Gene expression patterns that support novel developmental stress buffering in embryos of the annual killifish austrofundulus limnaeus. Evo Devo 2015, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Rogers, C.D.; Archer, T.C.; Cunningham, D.D.; Grammer, T.C.; Casey, E.M. Sox3 expression is maintained by FGF signaling and restricted to the neural plate by vent proteins in the xenopus embryo. Dev. Biol. 2008, 313, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Uwanogho, D.; Rex, M.; Cartwright, E.J.; Pearl, G.; Healy, C.; Scotting, P.J.; Sharpe, P.T. Embryonic expression of the chicken sox2, sox3 and soxll suggests an interactive role in neuronal development. Mech. Dev. 1995, 49, 23–36. [Google Scholar] [CrossRef]
- Rizzoti, K.; Brunelli, S.; Carmignac, D.; Thomas, P.Q.; Robinson, I.C.; Lovell-Badge, R. Sox3 is required during the formation of the hypothalamo-pituitary axis. Nat. Genet. 2004, 36, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Temple, S. The development of neural stem cells. Nature 2001, 414, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.S.; An, K.W.; Park, M.S.; Jeong, M.H.; Choi, C.Y. Quantitative mRNA expression of sox3 and DMRT1 during sex reversal, and expression profiles after GnRHa administration in black porgy, acanthopagrus schlegeli. Comp. Biochem. Physiol. B 2009, 154, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, J.; You, F.; Ma, L.; Yang, X.; Gao, J.; He, Y.; Qi, J.; Yu, H.; Wang, Z.; et al. Detection of alternative splice and gene duplication by RNA sequencing in japanese flounder, paralichthys olivaceus. Gene Genom. Genet. 2014, 4, 2419–2424. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information. Available online: http://www.ncbi.nlm.gov (accessed on 26 August 2015).
- ClustalW2. Available online: http://www.ebi.ac.uk/tools/msa/clustalw2/ (accessed on 26 August 2015).
- Simple Modular Architecture Research Tool. Available online: http://smart.embl-heidelberg.de/ (accessed on 26 August 2015).
- Interproscan. Available online: http://www.ebi.ac.uk/tools/pfa/iprscan/ (accessed on 26 August 2015).
- Matinspector. Available online: http://www.genomatix.de/matinspector.html (assessed on 26 August 2015).
- mVISTA. Available online: http://genome.Lbl.Gov/vista/mvista/submit.shtml (assessed on 26 August 2015).
- Meme Suite. Available online: http://meme-suite.org/tools/meme (assessed on 26 August 2015).
- Zhong, Q.; Zhang, Q.; Wang, Z.; Qi, J.; Chen, Y.; Li, S.; Sun, Y.; Li, C.; Lan, X. Expression profiling and validation of potential reference genes during paralichthys olivaceus embryogenesis. Mar. Biotechnol. 2008, 10, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hu, Y.H.; Sun, B.G.; Xiao, Z.Z.; Sun, L. Selection of normalization factors for quantitative real time RT-PCR studies in japanese flounder (paralichthys olivaceus) and turbot (scophthalmus maximus) under conditions of viral infection. Vet. Immunol. Immunopathol. 2013, 152, 303–316. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.-F.; Zhang, Z.-F.; Shao, M.-Y.; Zhu, W. Developmental expression pattern of the Fc-vasa-like gene, gonadogenesis and development of germ cell in chinese shrimp, fenneropenaeus chinensis. Aquaculture 2011, 314, 202–209. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, J.; Li, P.; Zhang, W.; Wang, Z.; Wang, X.; Zhang, Q. Molecular Cloning, Promoter Analysis and Expression Profiles of the sox3 Gene in Japanese Flounder, Paralichthys olivaceus. Int. J. Mol. Sci. 2015, 16, 27931-27944. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms161126079
Gao J, Li P, Zhang W, Wang Z, Wang X, Zhang Q. Molecular Cloning, Promoter Analysis and Expression Profiles of the sox3 Gene in Japanese Flounder, Paralichthys olivaceus. International Journal of Molecular Sciences. 2015; 16(11):27931-27944. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms161126079
Chicago/Turabian StyleGao, Jinning, Peizhen Li, Wei Zhang, Zhigang Wang, Xubo Wang, and Quanqi Zhang. 2015. "Molecular Cloning, Promoter Analysis and Expression Profiles of the sox3 Gene in Japanese Flounder, Paralichthys olivaceus" International Journal of Molecular Sciences 16, no. 11: 27931-27944. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms161126079