Plant Adaptation to Multiple Stresses during Submergence and Following Desubmergence
Abstract
:1. Introduction

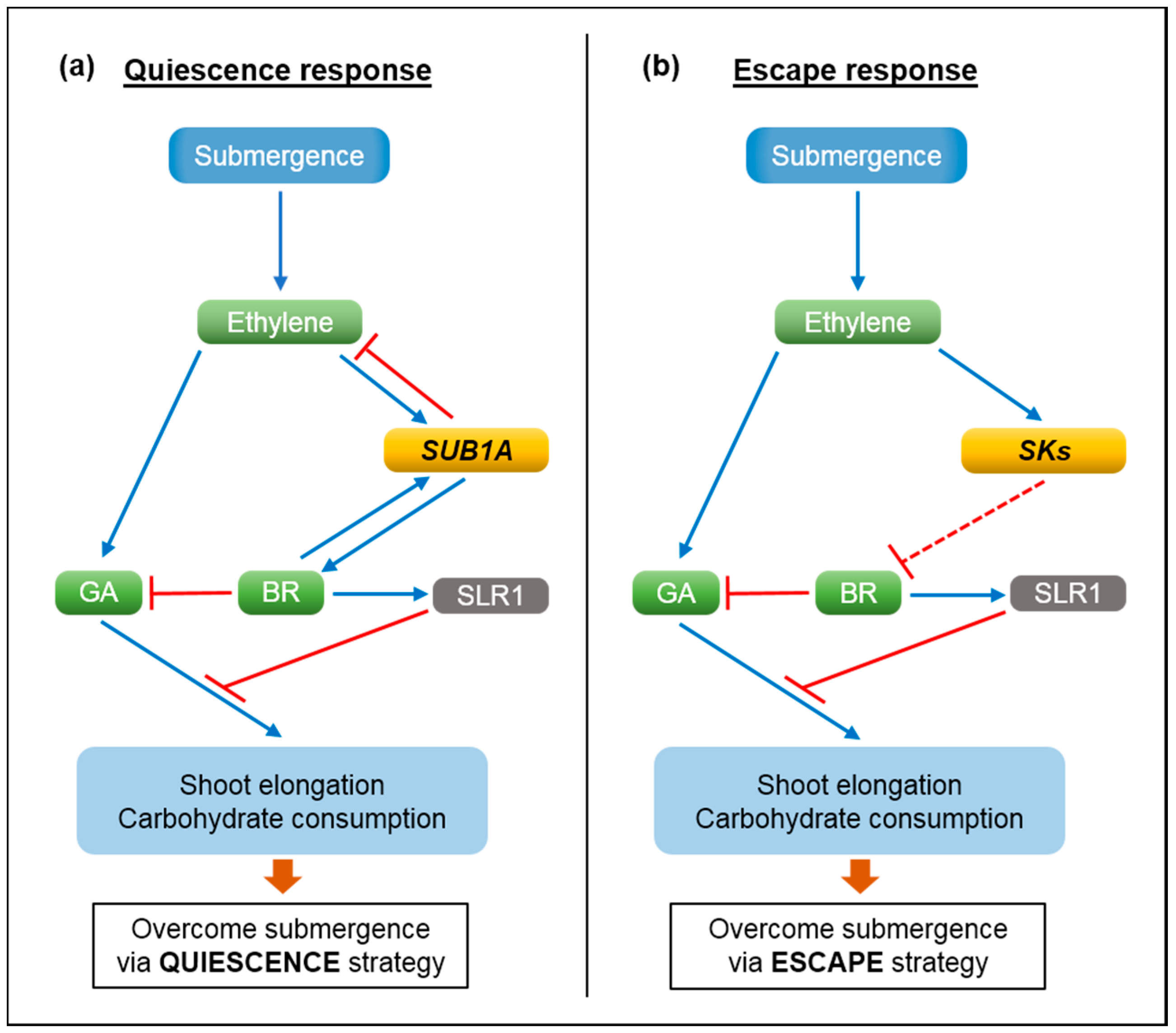
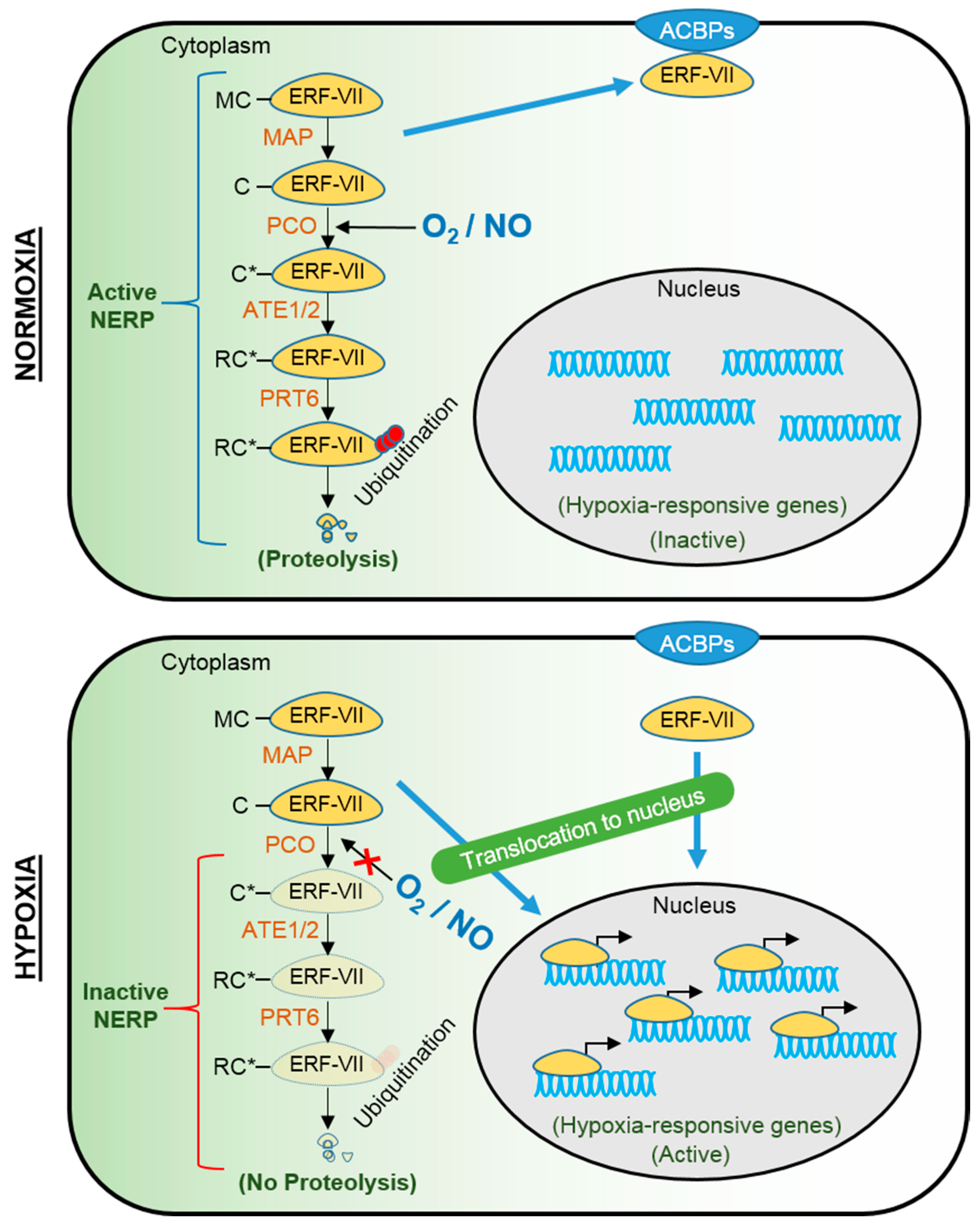
2. Molecular Mechanisms of Submergence Tolerance and Escape
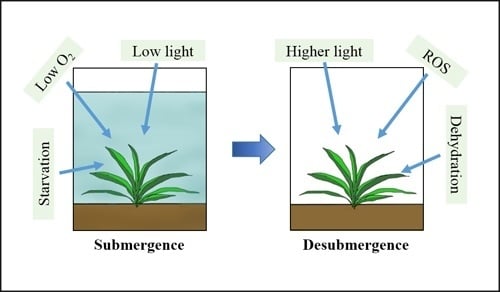
3. Submergence, Reoxygenation, and Dehydration
4. Submergence and Starvation

5. Submergence and Disease
6. Submergence and Salinity
7. Conclusions and Future Perspectives
| Gene | Species | Function | Tolerance | References |
|---|---|---|---|---|
| SUB1A | Rice | ERF-VII TF | Submergence a, oxidative stress a, drought a, prolonged darkness (starvation) a | [8,18,51] |
| EREBP1 | Rice | ERF-VII TF | Submergence a, drought a, disease a | [46] |
| SNORKEL1/2 | Rice | ERF-VII TF | Submergence (escape response) a | [20] |
| RAP2.2 | Arabidopsis | ERF-VII TF | Submergence a, low oxygen a, oxidative stress a, osmotic stress a, disease a | [27,29,90] |
| RAP2.3 | Arabidopsis | ERF-VII TF | Submergence a, low oxygen a, oxidative stress a, osmotic stress a | [27] |
| RAP2.12 | Arabidopsis | ERF-VII TF | Submergence a, low oxygen a, oxidative stress a, osmotic stress a | [27,30] |
| HRE1 | Arabidopsis | ERF-VII TF | Submergence a, low oxygen a | [26,32,34] |
| HRE2 | Arabidopsis | ERF-VII TF | Submergence a, low oxygen a, oxidative stress a, osmotic stress a | [26,31,34] |
| PCO1/2 | Arabidopsis | Cysteine oxidase | Submergence b | [38] |
| ATE1/2 | Arabidopsis | Arginine transferase | Low oxygen b | [34] |
| PRT6 | Arabidopsis | Ubiquitin ligase | Submergence b, low oxygen b, prolonged darkness (starvation) b | [34,40] |
| PRT6 | Barley | Ubiquitin ligase | Waterlogging b | [41] |
| CIPK15 | Rice | CBL-interacting protein kinase | Submergence (germination and early vegetative stage) a | [55] |
| SnRK1A | Rice | SNF1-related protein kinase | Submergence (germination and early vegetative stage) a | [55] |
| KIN10 | Arabidopsis | SNF1-related protein kinase | Submergence (early vegetative stage) a, senescence a, salinity b | [67,98,99] |
| TPP7 | Rice | T6P phosphatase | Submergence (germination and early vegetative stage) a | [59] |
| WRKY76 | Sunflower | WRKY TF | Submergence a, waterlogging a, drought a | [47] |
| WRKY22 | Arabidopsis | WRKY TF | Disease a | [89] |
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bailey-Serres, J.; Fukao, T.; Gibbs, D.J.; Holdsworth, M.J.; Lee, S.C.; Licausi, F.; Perata, P.; Voesenek, L.A.C.J.; van Dongen, J.T. Making sense of low oxygen sensing. Trends Plant. Sci. 2012, 17, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Voesenek, L.A.C.J.; Bailey-Serres, J. Flood adaptive traits and processes: An overview. New Phytol. 2015, 206, 57–73. [Google Scholar] [CrossRef] [PubMed]
- Bailey-Serres, J.; Voesenek, L.A.C.J. Flooding stress: Acclimations and genetic diversity. Annu. Rev. Plant. Biol. 2008, 59, 313–339. [Google Scholar] [CrossRef] [PubMed]
- Magneschi, L.; Perata, P. Rice germination and seedling growth in the absence of oxygen. Ann. Bot. 2009, 103, 181–196. [Google Scholar] [CrossRef] [PubMed]
- Bailey-Serres, J.; Colmer, T.D. Plant tolerance of flooding stress—recent advances. Plant Cell Environ. 2014, 37, 2211–2215. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, O. Long-distance water transport in aquatic plants. Plant Physiol. 1993, 103, 1369–1375. [Google Scholar] [PubMed]
- Crawford, R.M.M. Oxygen availability as an ecological limit to plant distribution. Adv. Ecol. Res. 1992, 23, 93–185. [Google Scholar]
- Fukao, T.; Yeung, E.; Bailey-Serres, J. The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant. Cell 2011, 23, 412–427. [Google Scholar] [CrossRef] [PubMed]
- Ruban, A.V. Evolution under the sun: Optimizing light harvesting in photosynthesis. J. Exp. Bot. 2015, 66, 7–23. [Google Scholar] [CrossRef] [PubMed]
- Setter, T.L.; Bhekasut, P.; Greenway, H. Desiccation of leaves after de-submergence is one cause for intolerance to complete submergence of the rice cultivar IR 42. Funct. Plant. Biol. 2010, 37, 1096–1104. [Google Scholar] [CrossRef]
- Harvell, C.D.; Mitchell, C.E.; Ward, J.R.; Altizer, S.; Dobson, A.P.; Ostfeld, R.S.; Samuel, M.D. Climate warming and disease risks for terrestrial and marine biota. Science 2002, 296, 2158–2162. [Google Scholar] [CrossRef] [PubMed]
- Melotto, M.; Underwood, W.; He, S.Y. Role of stomata in plant innate immunity and foliar bacterial diseases. Annu. Rev. Phytopathol. 2008, 46, 101–122. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Syvertsen, J.P.; McCoy, C.W.; Stuart, R.J.; Schumann, A.W. Water stress and root injury from simulated flooding and Diaprepes abbreviatus root weevil larval feeding in citrus. Soil Sci. 2006, 171, 138–151. [Google Scholar] [CrossRef]
- Erb, M.; Lu, J. Soil abiotic factors influence interactions between belowground herbivores and plant roots. J. Exp. Bot. 2013, 64, 1295–1303. [Google Scholar] [PubMed]
- Xu, K.; Xu, X.; Fukao, T.; Canlas, P.; Maghirang-Rodriguez, R.; Heuer, S.; Ismail, A.M.; Bailey-Serres, J.; Ronald, P.C.; Mackill, D.J. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 2006, 442, 705–708. [Google Scholar] [PubMed]
- Fukao, T.; Xiong, L. Genetic mechanisms conferring adaptation to submergence and drought in rice: Simple or complex? Curr. Opin. Plant Biol. 2013, 16, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Dang, T.T.M.; Vergara, G.V.; Pandey, D.M.; Sanchez, D.; Neeraja, C.N.; Septiningsih, E.M.; Mendioro, M.; Tecson-Mendoza, E.M.; et al. Molecular marker survey and expression analyses of the rice submergence-tolerance gene SUB1A. Theor. Appl. Genet. 2010, 121, 1441–1453. [Google Scholar] [CrossRef] [PubMed]
- Fukao, T.; Xu, K.; Ronald, P.C.; Bailey-Serres, J. A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 2006, 18, 2021–2034. [Google Scholar] [CrossRef] [PubMed]
- Fukao, T.; Bailey-Serres, J. Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc. Natl. Acad. Sci. USA 2008, 105, 16814–16819. [Google Scholar] [CrossRef] [PubMed]
- Hattori, Y.; Nagai, K.; Furukawa, S.; Song, X.J.; Kawano, R.; Sakakibara, H.; Wu, J.; Matsumoto, T.; Yoshimura, A.; Kitano, H.; et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 2009, 460, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, A.J.; Folsom, J.J.; Jikamaru, Y.; Ronald, P.; Walia, H. SUB1A-mediated submergence tolerance response in rice involves differential regulation of the brassinosteroid pathway. New Phytol. 2013, 198, 1060–1070. [Google Scholar] [PubMed]
- Ayano, M.; Kani, T.; Kojima, M.; Sakakibara, H.; Kitaoka, T.; Kuroha, T.; Angeles-Shim, R.B.; Kitano, H.; Nagai, K.; Ashikari, M. Gibberellin biosynthesis and signal transduction is essential for internode elongation in deepwater rice. Plant Cell Environ. 2014, 37, 2313–2324. [Google Scholar] [CrossRef] [PubMed]
- Kende, H.; van der Knaap, E.; Cho, H.-T. Deepwater rice: A model plant to study stem elongation. Plant Physiol. 1998, 118, 1105–1110. [Google Scholar] [CrossRef] [PubMed]
- Fukao, T.; Bailey-Serres, J. Ethylene—a key regulator of submergence responses in rice. Plant Sci. 2008, 175, 43–51. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; van Dongen, J.T.; Giuntoli, B.; Novi, G.; Santaniello, A.; Geigenberger, P.; Perata, P. HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J. 2010, 62, 302–315. [Google Scholar] [CrossRef] [PubMed]
- Papdi, C.; Perez-Salamo, I.; Joseph, M.P.; Giuntoli, B.; Bogre, L.; Koncz, C.; Szabados, L. The low oxygen, oxidative and osmotic stress responses synergistically act through the ethylene response factor VII genes RAP2.12, RAP2.2 and RAP2.3. Plant J. 2015, 82, 772–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mustroph, A.; Zanetti, M.E.; Jang, C.J.H.; Holtan, H.E.; Repetti, P.P.; Galbraith, D.W.; Girke, T.; Bailey-Serres, J. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 18843–18848. [Google Scholar] [CrossRef] [PubMed]
- Hinz, M.; Wilson, I.W.; Yang, J.; Buerstenbinder, K.; Llewellyn, D.; Dennis, E.S.; Sauter, M.; Dolferus, R. Arabidopsis RAP2.2: An ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 2010, 153, 757–772. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; Kosmacz, M.; Weits, D.A.; Giuntoli, B.; Giorgi, F.M.; Voesenek, L.A.C.J.; Perata, P.; van Dongen, J.T. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 2011, 479, 419–422. [Google Scholar] [CrossRef] [PubMed]
- Park, H.Y.; Seok, H.Y.; Woo, D.H.; Lee, S.Y.; Tarte, V.N.; Lee, E.H.; Lee, C.H.; Moon, Y.H. AtERF71/HRE2 transcription factor mediates osmotic stress response as well as hypoxia response in Arabidopsis. Biochem. Biophys. Res. Commun. 2011, 414, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Seok, H.Y.; Tarte, V.N.; Lee, S.Y.; Park, H.Y.; Moon, Y.H. Arabidopsis HRE1α, a splicing variant of AtERF73/HRE1, functions as a nuclear transcription activator in hypoxia response and root development. Plant Cell Rep. 2014, 33, 1255–1262. [Google Scholar] [CrossRef] [PubMed]
- Giuntoli, B.; Lee, S.C.; Licausi, F.; Kosmacz, M.; Oosumi, T.; van Dongen, J.T.; Bailey-Serres, J.; Perata, P. A trihelix DNA binding protein counterbalances hypoxia-responsive transcriptional activation in Arabidopsis. PLoS Biol. 2014, 12, e1001950. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, D.J.; Lee, S.C.; Isa, N.M.; Gramuglia, S.; Fukao, T.; Bassel, G.W.; Correia, C.S.; Corbineau, F.; Theodoulou, F.L.; Bailey-Serres, J.; et al. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 2011, 479, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, D.J.; Conde, J.V.; Berckhan, S.; Prasad, G.; Mendiondo, G.M.; Holdsworth, M.J. Group VII Ethylene Response Factors coordinate oxygen and nitric oxide signal transduction and stress responses in plants. Plant Physiol. 2015, 169, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, D.J.; Bacardit, J.; Bachmair, A.; Holdsworth, M.J. The eukaryotic N-end rule pathway: Conserved mechanisms and diverse functions. Trends Cell Biol. 2014, 24, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, D.J.; Md Isa, N.; Movahedi, M.; Lozano-Juste, J.; Mendiondo, G.M.; Berckhan, S.; Marin-de la Rosa, N.; Vicente Conde, J.; Sousa Correia, C.; Pearce, S.P.; et al. Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol. Cell 2014, 53, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Weits, D.A.; Giuntoli, B.; Kosmacz, M.; Parlanti, S.; Hubberten, H.M.; Riegler, H.; Hoefgen, R.; Perata, P.; van Dongen, J.T.; Licausi, F. Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat. Commun. 2014, 5, 3425. [Google Scholar] [CrossRef] [PubMed]
- Kosmacz, M.; Parlanti, S.; Schwarzlander, M.; Kragler, F.; Licausi, F.; Van Dongen, J.T. The stability and nuclear localization of the transcription factor RAP2.12 are dynamically regulated by oxygen concentration. Plant Cell Environ. 2015, 38, 1094–1103. [Google Scholar] [CrossRef] [PubMed]
- Riber, W.; Muller, J.T.; Visser, E.J.; Sasidharan, R.; Voesenek, L.A.C.J.; Mustroph, A. The Greening after Extended Darkness1 is an N-end rule pathway mutant with high tolerance to submergence and starvation. Plant Physiol. 2015, 167, 1616–1629. [Google Scholar] [CrossRef] [PubMed]
- Mendiondo, G.M.; Gibbs, D.J.; Szurman-Zubrzycka, M.; Korn, A.; Marquez, J.; Szarejko, I.; Maluszynski, M.; King, J.; Axcell, B.; Smart, K.; et al. Enhanced waterlogging tolerance in barley by manipulation of expression of the N-end rule pathway E3 ligase PROTEOLYSIS6. Plant Biotechnol. J. 2015. [Google Scholar] [CrossRef]
- Blokhina, O.; Virolainen, E.; Fagerstedt, K.V. Antioxidants, oxidative damage and oxygen deprivation stress: a review. Ann. Bot. 2003, 91, 179–194. [Google Scholar] [CrossRef] [PubMed]
- Boamfa, E.I.; Ram, P.C.; Jackson, M.B.; Reuss, J.; Harren, F.J.M. Dynamic aspects of alcoholic fermentation of rice seedlings in response to anaerobiosis and to complete submergence: Relationship to submergence tolerance. Ann. Bot. 2003, 91, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Boamfa, E.I.; Veres, A.H.; Ram, P.C.; Jackson, M.B.; Reuss, J.; Harren, F.J. Kinetics of ethanol and acetaldehyde release suggest a role for acetaldehyde production in tolerance of rice seedlings to micro-aerobic conditions. Ann. Bot. 2005, 96, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Alpuerto, J.B.; Hussain, R.M.F.; Fukao, T. The key regulator of submergence tolerance, SUB1A, promotes photosynthetic and metabolic recovery from submergence damage in rice leaves. Plant Cell Environ. 2015. [Google Scholar] [CrossRef] [PubMed]
- Jisha, V.; Dampanaboina, L.; Vadassery, J.; Mithofer, A.; Kappara, S.; Ramanan, R. Overexpression of an AP2/ERF type transcription factor OsEREBP1 confers biotic and abiotic stress tolerance in rice. PLoS ONE 2015, 10, e0127831. [Google Scholar] [CrossRef] [PubMed]
- Raineri, J.; Ribichich, K.F.; Chan, R.L. The sunflower transcription factor HaWRKY76 confers drought and flood tolerance to Arabidopsis thaliana plants without yield penalty. Plant Cell Rep. 2015, 34, 2065–2080. [Google Scholar] [CrossRef] [PubMed]
- Colmer, T.D.; Pedersen, O. Underwater photosynthesis and respiration in leaves of submerged wetland plants: gas films improve CO2 and O2 exchange. New Phytol. 2008, 177, 918–926. [Google Scholar] [CrossRef] [PubMed]
- Winkel, A.; Pedersen, O.; Ella, E.; Ismail, A.M.; Colmer, T.D. Gas film retention and underwater photosynthesis during field submergence of four contrasting rice genotypes. J. Exp. Bot. 2014, 65, 3225–3233. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M.B.; Ram, P.C. Physiological and molecular basis of susceptibility and tolerance of rice plants to complete submergence. Ann. Bot. 2003, 91, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Fukao, T.; Yeung, E.; Bailey-Serres, J. The submergence tolerance gene SUB1A delays leaf senescence under prolonged darkness through hormonal regulation in rice. Plant Physiol. 2012, 160, 1795–1807. [Google Scholar] [CrossRef] [PubMed]
- Miro, B.; Ismail, A.M. Tolerance of anaerobic conditions caused by flooding during germination and early growth in rice (Oryza sativa L.). Front. Plant Sci. 2013, 4, 269. [Google Scholar] [CrossRef] [PubMed]
- Ismail, A.M.; Ella, E.S.; Vergara, G.V.; Mackill, D.J. Mechanisms associated with tolerance to flooding during germination and early seedling growth in rice (Oryza sativa). Ann. Bot. 2009, 103, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.-A.; Lin, C.-C.; Lee, K.-W.; Chen, J.-L.; Huang, L.-F.; Ho, S.-L.; Liu, H.-J.; Hsing, Y.-I.; Yu, S.-M. The SnRK1A protein kinase plays a key role in sugar signaling during germination and seedling growth of rice. Plant Cell 2007, 19, 2484–2499. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-W.; Chen, P.-W.; Lu, C.-A.; Chen, S.; Ho, T.-H.D.; Yu, S.-M. Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci. Signal. 2009, 2. [Google Scholar] [CrossRef] [PubMed]
- Polge, C.; Thomas, M. SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 2007, 12, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Angaji, S.A.; Septiningsih, E.M.; Mackill, D.J.; Ismail, A.M. QTLs associated with tolerance of flooding during germination in rice (Oryza sativa L.). Euphytica 2010, 172, 159–168. [Google Scholar] [CrossRef]
- Septiningsih, E.M.; Ignacio, J.C.; Sendon, P.M.; Sanchez, D.L.; Ismail, A.M.; Mackill, D.J. QTL mapping and confirmation for tolerance of anaerobic conditions during germination derived from the rice landrace Ma-Zhan Red. Theor. Appl. Genet. 2013, 126, 1357–1366. [Google Scholar] [CrossRef] [PubMed]
- Kretzschmar, T.; Pelayo, M.A.F.; Trijatmiko, K.R.; Gabunada, L.F.M.; Alam, R.; Jimenez, R.; Mendioro, M.S.; Slamet-Loedin, I.H.; Sreenivasulu, N.; Bailey-Serres, J.; et al. A trehalose-6-phosphate phosphatase enhances anaerobic germination tolerance in rice. Nat. Plants 2015, 1, 15124. [Google Scholar] [CrossRef]
- Zhang, Y.; Primavesi, L.F.; Jhurreea, D.; Andralojc, P.J.; Mitchell, R.A.; Powers, S.J.; Schluepmann, H.; Delatte, T.; Wingler, A.; Paul, M.J. Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol. 2009, 149, 1860–1871. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.J.; Jhurreea, D.; Zhang, Y.; Primavesi, L.F.; Delatte, T.; Schluepmann, H.; Wingler, A. Upregulation of biosynthetic processes associated with growth by trehalose 6-phosphate. Plant Signal. Behav. 2010, 5, 386–392. [Google Scholar] [CrossRef] [PubMed]
- Delatte, T.L.; Sedijani, P.; Kondou, Y.; Matsui, M.; de Jong, G.J.; Somsen, G.W.; Wiese-Klinkenberg, A.; Primavesi, L.F.; Paul, M.J.; Schluepmann, H. Growth arrest by trehalose-6-phosphate: An astonishing case of primary metabolite control over growth by way of the SnRK1 signaling pathway. Plant Physiol. 2011, 157, 160–174. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shewry, P.R.; Jones, H.; Barcelo, P.; Lazzeri, P.A.; Halford, N.G. Expression of antisense SnRK1 protein kinase sequence causes abnormal pollen development and male sterility in transgenic barley. Plant J. 2001, 28, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Lovas, Á.; Bimbó, A.; Szabó, L.; Bánfalvi, Z. Antisense repression of StubGAL83 affects root and tuber development in potato. Plant J. 2003, 33, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Satoh-Nagasawa, N.; Nagasawa, N.; Malcomber, S.; Sakai, H.; Jackson, D. A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 2006, 441, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Van Dijken, A.J.; Schluepmann, H.; Smeekens, S.C. Arabidopsis trehalose-6-phosphate synthase 1 is essential for normal vegetative growth and transition to flowering. Plant Physiol. 2004, 135, 969–977. [Google Scholar] [CrossRef] [PubMed]
- Baena-Gonzalez, E.; Rolland, F.; Thevelein, J.M.; Sheen, J. A central integrator of transcription networks in plant stress and energy signalling. Nature 2007, 448, 938–942. [Google Scholar] [CrossRef] [PubMed]
- Tsai, A.Y.-L.; Gazzarrini, S. AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J. 2012, 69, 809–821. [Google Scholar] [CrossRef] [PubMed]
- Wahl, V.; Ponnu, J.; Schlereth, A.; Arrivault, S.; Langenecker, T.; Franke, A.; Feil, R.; Lunn, J.E.; Stitt, M.; Schmid, M. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 2013, 339, 704–707. [Google Scholar] [CrossRef] [PubMed]
- Amtmann, A.; Troufflard, S.; Armengaud, P. The effect of potassium nutrition on pest and disease resistance in plants. Physiol. Plant. 2008, 133, 682–691. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Blumwald, E. Genetic engineering for modern agriculture: Challenges and perspectives. Annu. Rev. Plant Biol. 2010, 61, 443–462. [Google Scholar] [CrossRef] [PubMed]
- Koga, H.; Dohi, K.; Mori, M. Abscisic acid and low temperatures suppress the whole plant-specific resistance reaction of rice plants to the infection of Magnaporthe grisea. Physiol. Mol. Plant Pathol. 2004, 65, 3–9. [Google Scholar] [CrossRef]
- Mohr, P.G.; Cahill, D.M. Abscisic acid influences the susceptibility of Arabidopsis thaliana to Pseudomonas syringae pv. tomato and Peronospora. parasitica. Funct. Plant Biol. 2003, 30, 461–469. [Google Scholar] [CrossRef]
- Kunkel, B.N.; Brooks, D.M. Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 2002, 5, 325–331. [Google Scholar] [CrossRef]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D.G. Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef] [PubMed]
- Audenaert, K.; De Meyer, G.B.; Hofte, M.M. Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol. 2002, 128, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.P.; Badruzsaufari, E.; Schenk, P.M.; Manners, J.M.; Desmond, O.J.; Ehlert, C.; Maclean, D.J.; Ebert, P.R.; Kazan, K. Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 2004, 16, 3460–3479. [Google Scholar] [CrossRef] [PubMed]
- Mauch-Mani, B.; Mauch, F. The role of abscisic acid in plant-pathogen interactions. Curr. Opin. Plant Biol. 2005, 8, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Fujita, Y.; Noutoshi, Y.; Takahashi, F.; Narusaka, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Crosstalk between abiotic and biotic stress responses: A current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 2006, 9, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, M.; Ishikawa, A.; Jikumaru, Y.; Seki, M.; Umezawa, T.; Asami, T.; Maruyama-Nakashita, A.; Kudo, T.; Shinozaki, K.; Yoshida, S.; et al. Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis. Plant Cell 2008, 20, 1678–1692. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, O.; Chico, J.M.; Sanchez-Serrano, J.J.; Solano, R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 2004, 16, 1938–1950. [Google Scholar] [CrossRef] [PubMed]
- Khong, G.N.; Pati, P.K.; Richaud, F.; Parizot, B.; Bidzinski, P.; Mai, C.D.; Bes, M.; Bourrie, I.; Meynard, D.; Beeckman, T.; et al. OsMADS26 negatively regulates resistance to pathogens and drought tolerance in rice. Plant Physiol. 2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Huang, L.; Hong, Y.; Zhang, Y.; Liu, S.; Li, D.; Zhang, H.; Song, F. Co-silencing of tomato S-adenosylhomocysteine hydrolase genes confers increased immunity against Pseudomonas syringae pv. tomato DC3000 and enhanced tolerance to drought stress. Front. Plant Sci. 2015, 6, 717. [Google Scholar] [CrossRef] [PubMed]
- Hammond-Kosack, K.E.; Silverman, P.; Raskin, I.; Jones, J.D.G. Race-specific elicitors of Cladosporium. fulvum induce changes in cell morphology and the synthesis of ethylene and salicylic acid in tomato plants carrying the corresponding Cf. disease resistance gene. Plant Physiol. 1996, 110, 1381–1394. [Google Scholar] [PubMed]
- Wang, C.; Cai, X.; Zheng, Z. High humidity represses Cf-4/Avr4- and Cf-9/Avr9-dependent hypersensitive cell death and defense gene expression. Planta 2005, 222, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Jambunathan, N.; Siani, J.M.; McNellis, T.W. A humidity-sensitive Arabidopsis copine mutant exhibits precocious cell death and increased disease resistance. Plant Cell 2001, 13, 2225–2240. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, K.; Kachroo, P.; Tsui, F.; Sharma, S.B.; Shah, J.; Klessig, D.F. Environmentally sensitive, SA-dependent defense responses in the cpr22 mutant of Arabidopsis. Plant J. 2001, 26, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Menke, F.L.; Yoshioka, K.; Moder, W.; Shirano, Y.; Klessig, D.F. High humidity suppresses ssi4-mediated cell death and disease resistance upstream of MAP kinase activation, H2O2 production and defense gene expression. Plant J. 2004, 39, 920–932. [Google Scholar] [CrossRef] [PubMed]
- Hsu, F.-C.; Chou, M.-Y.; Chou, S.-J.; Li, Y.-R.; Peng, H.-P.; Shih, M.-C. Submergence confers immunity mediated by the WRKY22 transcription factor in Arabidopsis. Plant Cell 2013, 25, 2699–2713. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wei, T.; Yin, K.Q.; Chen, Z.; Gu, H.; Qu, L.J.; Qin, G. Arabidopsis RAP2.2 plays an important role in plant resistance to Botrytis cinerea and ethylene responses. New Phytol. 2012, 195, 450–60. [Google Scholar] [CrossRef] [PubMed]
- Colmer, T.D.; Flowers, T.J. Flooding tolerance in halophytes. New Phytol. 2008, 179, 964–974. [Google Scholar] [CrossRef] [PubMed]
- Gierth, M.; Maser, P. Potassium transporters in plants—involvement in K+ acquisition, redistribution and homeostasis. FEBS Lett. 2007, 581, 2348–2356. [Google Scholar] [CrossRef] [PubMed]
- Plett, D.C.; Moller, I.S. Na+ transport in glycophytic plants: What we know and would like to know. Plant Cell Environ. 2010, 33, 612–626. [Google Scholar] [CrossRef] [PubMed]
- Teakle, N.L.; Tyerman, S.D. Mechanisms of Cl− transport contributing to salt tolerance. Plant Cell Environ. 2010, 33, 566–589. [Google Scholar] [CrossRef] [PubMed]
- Barrett-Lennard, E.G.; Shabala, S.N. The waterlogging/salinity interaction in higher plants revisited—focusing on the hypoxia-induced disturbance to K+ homeostasis. Funct. Plant Biol. 2013, 40, 872–882. [Google Scholar] [CrossRef]
- Kotula, L.; Clode, P.L.; Striker, G.G.; Pedersen, O.; Lauchli, A.; Shabala, S.; Colmer, T.D. Oxygen deficiency and salinity affect cell-specific ion concentrations in adventitious roots of barley (Hordeum vulgare). New Phytol. 2015, 208, 1114–1125. [Google Scholar] [CrossRef] [PubMed]
- Teakle, N.L.; Colmer, T.D.; Pedersen, O. Leaf gas films delay salt entry and enhance underwater photosynthesis and internal aeration of Melilotus siculus submerged in saline water. Plant Cell Environ. 2014, 37, 2339–2349. [Google Scholar] [PubMed]
- Im, J.H.; Cho, Y.H.; Kim, G.D.; Kang, G.H.; Hong, J.W.; Yoo, S.D. Inverse modulation of the energy sensor Snf1-related protein kinase 1 on hypoxia adaptation and salt stress tolerance in Arabidopsis thaliana. Plant Cell Environ. 2014, 37, 2303–2312. [Google Scholar] [PubMed]
- Cho, Y.H.; Hong, J.W.; Kim, E.C.; Yoo, S.D. Regulatory functions of SnRK1 in stress-responsive gene expression and in plant growth and development. Plant Physiol. 2012, 158, 1955–1964. [Google Scholar] [CrossRef] [PubMed]
- Fukao, T.; Harris, T.; Bailey-Serres, J. Evolutionary analysis of the Sub1 gene cluster that confers submergence tolerance to domesticated rice. Ann. Bot. 2009, 103, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Setter, T.L.; Waters, I.; Sharma, S.K.; Singh, K.N.; Kulshreshtha, N.; Yaduvanshi, N.P.; Ram, P.C.; Singh, B.N.; Rane, J.; McDonald, G.; et al. Review of wheat improvement for waterlogging tolerance in Australia and India: The importance of anaerobiosis and element toxicities associated with different soils. Ann. Bot. 2009, 103, 221–235. [Google Scholar] [CrossRef] [PubMed]
- Pezeshki, S.R.; DeLaune, R.D. Soil oxidation-reduction in wetlands and its impact on plant functioning. Biology 2012, 1, 196–221. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Ito, Y.; Yamaguchi-Shinozaki, K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009, 149, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Holopainen, J.K.; Gershenzon, J. Multiple stress factors and the emission of plant VOCs. Trends Plant Sci. 2010, 15, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, P.Y.; Lerdau, M.T. Catabolism of volatile organic compounds influences plant survival. Trends Plant Sci. 2013, 18, 695–703. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tamang, B.G.; Fukao, T. Plant Adaptation to Multiple Stresses during Submergence and Following Desubmergence. Int. J. Mol. Sci. 2015, 16, 30164-30180. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms161226226
Tamang BG, Fukao T. Plant Adaptation to Multiple Stresses during Submergence and Following Desubmergence. International Journal of Molecular Sciences. 2015; 16(12):30164-30180. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms161226226
Chicago/Turabian StyleTamang, Bishal Gole, and Takeshi Fukao. 2015. "Plant Adaptation to Multiple Stresses during Submergence and Following Desubmergence" International Journal of Molecular Sciences 16, no. 12: 30164-30180. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms161226226