Fungal Community Successions in Rhizosphere Sediment of Seagrasses Enhalus acoroides under PAHs Stress
Abstract
:1. Introduction
2. Results and Discussion
2.1. Sediment Characteristics
| Sample | TP (%) | TN (%) | TC (%) | C/N Ratio |
|---|---|---|---|---|
| In situ Sediment | 0.0035 | 0.021 | 0.384 | 18.29 |
2.2. DGGE (Denaturing Gradient Gel Electrophoresis) Patterns the Fungal Communities under PAH (Polycyclic Aromatic Hydrocarbon) Stress

2.3. Dynamics of Shannon Index, Fungal Abundance Analysis

2.4. Clone Library Analysis

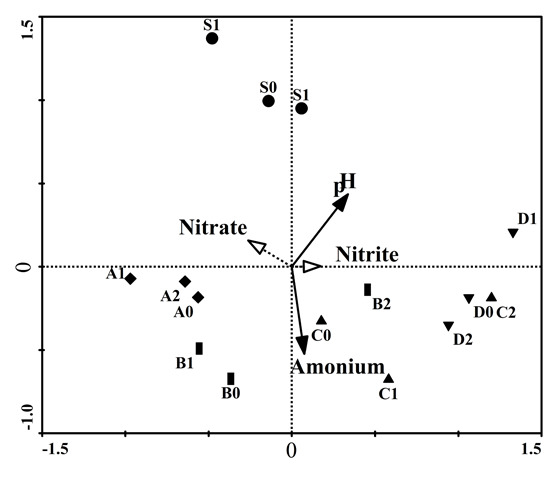
2.5. Redundancy Analysis, Variation Partitioning and Unweighted Pair Group Method with Arithmetic Mean (UMPGA)

| Axis | Eigen Value | Species-Environment Correlation | Cumulative % Variations of Species | Cumulative % Variations of Species-Environment | Sum of All Canonical Eigenvalue |
|---|---|---|---|---|---|
| - | - | - | - | - | 0.456 |
| Axis 1 | 0.279 | 0.864 | 27.9 | 61.3 | - |
| Axis 2 | 0.081 | 0.865 | 36.0 | 79.0 | - |
| Axis 3 | 0.062 | 0.790 | 42.2 | 92.6 | - |
| Axis 4 | 0.034 | 0.735 | 45.6 | 100.0 | - |
| Parameters Included in the Model | Eigen Value | Variation Explains Solely (%) | F-Value | p-Value |
|---|---|---|---|---|
| Ammonium | 0.173 | 37.90 | 3.010 | 0.004 |
| pH | 0.207 | 45.40 | 3.879 | 0.002 |
| All the above together | 0.311 | 68.20 | 2.097 | 0.004 |
3. Experimental Section
3.1. Sample Collection
3.2. Experimental Setup
3.3. DNA Extraction, Clone Library and PCR-DGGE
3.4. qPCR
3.5. Sequencing, Phylogenetic Analysis and Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hemminga, M.A.; Duarte, C.M. Seagrass Ecology, 1st ed.; Cambridge University Press: Cambridge, UK, 2000; p. 298. [Google Scholar]
- Short, F.T.; Polidoro, B.; Livingstone, S.R.; Carpenter, K.E.; Bandeira, S.; Bujang, J.S.; Calumpong, H.P.; Carruthers, T.J.B.; Coles, R.G.; Dennison, W.C.; et al. Extinction risk assessment of the world’s seagrass species. Biol. Conserv. 2011, 144, 1961–1971. [Google Scholar] [CrossRef] [Green Version]
- Lewis, M.A.; Dantin, D.D.; Chancy, C.A.; Abel, K.C.; Lewis, C.G. Florida seagrass habitat evaluation: A comparative survey for chemical quality. Environ. Pollut. 2007, 146, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Raja, S.; Thangaradjou, T.; Sivakumar, K.; Kannan, L. Rhizobacterial population density and nitrogen fixation in seagrass community of Gulf of Mannar, India. J. Environ. Biol. 2012, 33, 1033–1037. [Google Scholar] [PubMed]
- Heather, G. The microbial role in carbon cycling within seagrass sediments. Plymouth Stud. Sci. 2010, 3, 234–244. [Google Scholar]
- Pereg, L.L.; Lipkin, Y.; Sar, N. Different niches of the Halophila stipulacea seagrass bed harbour distinct populations of nitrogen fixing bacteria. Mar. Biol. 1994, 119, 327–333. [Google Scholar] [CrossRef]
- Waycott, M.; Duarte, C.M.; Carruthers, T.J.B.; Orth, R.J.; Dennison, W.C.; Olyarnik, S.; Calladine, A.; Fourqurean, J.W.; Heck, K.L., Jr.; Hughes, A.R.; et al. Accelerating loss of seagrasses across the globe threatens coastal ecosystems. Proc. Natl. Acad. Sci. USA 2009, 106, 12377–12381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zadeh, C.M.; Saify, A.; Shalikar, H. Polycyclic aromatic hydrocarbons (PAHs) along the eastern Caspian Sea coast. Glob. Environ. Res. 2010, 4, 59–63. [Google Scholar]
- Apostolopoulou, M.V.; Monteyne, E.; Krikonis, K.; Pavlopoulos, K.; Roose, P.; Dehairs, F. Monitoring polycyclic aromatic hydrocarbons in the Northeast Aegean Sea using Posidonia oceanica seagrass and synthetic passive samplers. Mar. Pollut. Bull. 2014, 87, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.H.; Pang, K.L.; Wu, Y.R.; Gu, J.D.; Chow, R.K.K.; Vrijmoed, L.L.P. Degradation of phthalate esters by Fusarium sp. DMT-5-3 and Trichosporon sp. DMl-5-1 isolated from mangrove sediments. In Biology of Marine Fungi, Progress in Molecular and Subcellular Biology; Raghukumar, C., Ed.; Springer-Verlag: Heidelberg, Germany, 2012; p. 53. [Google Scholar]
- Panno, L.; Bruno, M.; Voyron, S.; Anastasi, A.; Gnavi, G.; Miserere, L.; Varese, G.C. Diversity, ecological role and potential biotechnological applications of marine fungi associated to the seagrass Posidonia oceanic L. N. Biotechnol. 2013, 30, 685–694. [Google Scholar] [CrossRef] [PubMed]
- Zuccaro, A.; Schoch, C.L.; Spatafora, J.W.; Kohlmeyer, J.; Draeger, S.; Mitchell, J.I. Detection and identification of fungi intimately associated with the brown seaweed Fucus serratus. Appl. Environ. Microbiol. 2008, 74, 931–941. [Google Scholar] [CrossRef] [PubMed]
- García-Rodríguez, D.; Carro-Díaz, A.M.; Lorenzo-Ferreira, R.A. Supercritical fluid extraction of polyhalogenated pollutants from aquaculture and marine environmental samples: A review. J. Sep. Sci. 2008, 31, 1333–1345. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.Y.; Chu, Y.L.; Gu, J.D. Distribution and sources of polycyclic aromatic hydrocarbons in sediments of the Mai Po Inner Deep Bay Ramsar Site in Hong Kong. Ecotoxicology 2012, 21, 1743–1752. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, G.; Li, X.D.; Qi, S.H.; Liu, G.Q.; Peng, X.Z. Source seasonality of polycyclic aromatic hydrocarbons (PAHs) in a subtropical city, Guangzhou, South China. Sci. Total Environ. 2006, 355, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Northcott, G.L.; Kevin, C.J. Partitioning, extractability and formation of nonextractable PAH residues in soil. 1. Compound differences in aging and sequestration. Environ. Sci. Technol. 2001, 35, 1103–1110. [Google Scholar] [CrossRef] [PubMed]
- Sakayaroj, J.; Preedanon, S.; Supaphon, O.; Jones, E.B.G.S. Phongpaichit Phylogenetic diversity of endophyte assemblages associated with the tropical seagrass Enhalus acoroides in Thailand. Fungal Divers. 2010, 42, 27–45. [Google Scholar] [CrossRef]
- Mineki, S.; Suzuki, K.; Iwata, K.; Nakajima, D.; Goto, S. Degradation of Polyaromatic Hydrocarbons by Fungi Isolated from Soil in Japan. Polycycl. Aromat. Compd. 2015, 35, 120–128. [Google Scholar] [CrossRef]
- Wu, Y.R.; Luo, Z.H.; Vrijmoed, L.L. Biodegradation of Anthracene and Benz[a]anthracene by Two Fusarium solani Strains Isolated from Mangrove Sediments. Bioresour. Technol. 2010, 101, 9666–9672. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, J. Detoxification of polycyclic aromatic hydrocarbons by fungi. J. Ind. Microbiol. 1992, 9, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Zhang, Y.; El-Latif Hesham, A.; Liu, R.; Yang, M. Cell surface properties of five polycyclic aromatic compound-degrading yeast strains. Appl. Microbiol. Biotechnol. 2010, 86, 1933–1939. [Google Scholar] [CrossRef] [PubMed]
- Salvo, V.S.; Gallizia, I.; Moreno, M.; Fabiano, M. Fungal communities in PAH-impacted sediments of Genoa-Voltri Harbour (NW Mediterranean, Italy). Mar. Pollut. Bull. 2005, 50, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Haimann, R.A. Fungal technologies for the treatment of hazardous waste. Environ. Prog. 1995, 14, 201–203. [Google Scholar] [CrossRef]
- Wilson, W.L. Isolation of Endophytes from Seagrasses from Bermuda. Master’s Thesis, The University of New Brunswick, Saint John, NB, Canada, 1998. [Google Scholar]
- Panno, L.; Voyron, S.; Anastasi, A.; Varese, G.C. Marine fungi associated with the seagrass Posidonia oceanic L.: A potential source of novel metabolites and enzymes. J. Biotechnol. 2010, 150 (Suppl. 1), 383–354. [Google Scholar] [CrossRef]
- Supaphon, P.; Phongpaichit, S.; Rukachaisirikul, V.; Sakayaroj, J. Antimicrobial potential of endophytic fungi derived from three seagrass species: Cymodocea serrulata, Halophila ovalis and Thalassia hemprichii. PLoS ONE 2013, 8, e72520. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.W.; Wong, A.H.Y.; Yu, R.M.K.; Park, Y.D.; Wong, Y.S.; Tam, N.F.Y. Polycyclic aromatic hydrocarbon-induced structural shift of bacterial communities in mangrove sediment. Microbiol. Ecol. 2008, 58, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, A.R.; Karlson, U. Diffuse PAH contamination of surface soils: Environmental occurrence, bioavailability, and microbial degradation. Appl. Microbiol. Biotechnol. 2007, 76, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Alias, S.A.; Zainuddin, N.; Jones, G.E.B. Biodiversity of marine fungi in Malaysian mangroves. Bot. Mar. 2010, 53, 545–554. [Google Scholar] [CrossRef]
- Covino, S.; D’Annibale, A.; Stazi, S.R.; Cajthaml, T.; Čvančarová, M.; Stella, T.; Petruccioli, M. Assessment of degradation potential of aliphatic hydrocarbons by autochthonous filamentous fungi from a historically polluted clay soil. Sci. Total Environ. 2015, 505, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Annette, E.; Reinhard, W.; Bernd, M. Fate and stability of nonextractable residues of PAH on contaminated soils under environmental stress conditions. Environ. Sci. Technol. 1998, 32, 2585–2590. [Google Scholar]
- Totsche, K.U.; Danzer., J.; Kogel-Knabner, I. Dissolved organic matter-enhanced retention of polycyclic aromatic hydrocarbons in soil miscible displacement experiments. J. Environ. Qual. 1997, 26, 1090–1100. [Google Scholar] [CrossRef]
- El-Latif Hesham, A.; Khan, S.; Liu, X.; Zhang, Y.; Wang, Z.; Yang, M. Application of PCR-DGGE to analyse the yeast population dynamics in slurry reactors during degradation of polycyclic aromatic hydrocarbons in weathered oil. Yeast 2006, 23, 879–887. [Google Scholar]
- Marusenko, Y.; Huber, D.P.; Hall, S.J. Fungi mediate nitrous oxide production but not ammonia oxidation in arid land soils of the southwestern US. Soil Biol. Biochem. 2013, 63, 24–36. [Google Scholar] [CrossRef]
- Riser-Roberts, E. Remediation of Petroleum Contaminated Soils: Biological, Physical, and Chemical Processes, 1st ed.; CRC Press: Boca Raton, FL, USA, 2010; pp. 229–230. [Google Scholar]
- Nielsen, J.; Nielsen, P.H.; Frisvad, J.C. Fungal depside, guisinol, from a marine derived strain of Emericella unguis. Phytochemistry 1999, 50, 263–265. [Google Scholar] [CrossRef]
- Jiang, Z.; Huang, X.; Zhang, J.; Zhou, C.; Lian, Z.; Ni, Z. The effects of air exposure on the desiccation rateand photosynthetic activity of Thalassia hemprichii and Enhalus acoroides. Mar. Biol. 2014, 161, 1051–1061. [Google Scholar] [CrossRef]
- Wu, P.; Wang, Y.S.; Sun, C.C.; Sun, F.L.; Wang, Y.T. Microbial community shift with decabromodiphenyl ether (BDE 209) in sediments of the Pearl River estuary, China. Biologia 2013, 68, 788–796. [Google Scholar] [CrossRef]
- May, L.A.; Smiley, B.; Schmidt, M.G. Comparative denaturing gradient gel electrophoresis analysis of fungal communities associated with whole plant corn silage. Can. J. Microbiol. 2001, 47, 829–841. [Google Scholar] [CrossRef] [PubMed]
- Rousk, J.; Baath, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evolution. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Roesti, D.; Gaur, R.; Johri, B.N.; Imfeld, G.; Sharma, S.; Kawaljeet, K.; Aragno, M. Plant growth stage, fertilizer management and bio-inoculation of arbuscular mycorrhizal fungi and plant growth promoting rhizobacteria affect the rhizobacterial community structure in rain-fed wheat fields. Soil Biol. Biochem. 2006, 38, 1111–1120. [Google Scholar] [CrossRef]
- Zhang, J.C.; Zeng, G.M.; Chen, Y.N.; Yu, M.; Yu, Z.; Li, H.; Yu, Y.; Huang, H. Effects of physico-chemical parameters on the bacterial and fungal communities during agricultural waste composting. Bioresour. Technol. 2011, 102, 2950–2956. [Google Scholar] [CrossRef] [PubMed]
- Lepš, J.; Šmilauer, P. Multivariate Analysis of Ecological Data Using CANOCO, 3rd ed.; Cambridge University Press: Cambridge, UK, 2003; pp. 43–75. [Google Scholar]
- Zhang, Y.Y.; Dong, J.D.; Yang, Z.H.; Zhang, S.; Wang, Y.S. Phylogenetic diversity of nitrogen-fixing bacteria in mangrove sediments assessed by PCR-denaturing gradient gel electrophoresis. Arch. Microbiol. 2008, 190, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Ling, J.; Dong, J.D.; Wang, Y.S.; Zhang, Y.Y.; Deng, C.; Lin, L.; Wu, M.L; Sun, F.L. Spatial variation of bacterial community structure of the Northern South China Sea in relation to water chemistry. Ecotoxicoloy 2012, 21, 1669–1679. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ling, J.; Zhang, Y.; Wu, M.; Wang, Y.; Dong, J.; Jiang, Y.; Yang, Q.; Zeng, S. Fungal Community Successions in Rhizosphere Sediment of Seagrasses Enhalus acoroides under PAHs Stress. Int. J. Mol. Sci. 2015, 16, 14039-14055. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms160614039
Ling J, Zhang Y, Wu M, Wang Y, Dong J, Jiang Y, Yang Q, Zeng S. Fungal Community Successions in Rhizosphere Sediment of Seagrasses Enhalus acoroides under PAHs Stress. International Journal of Molecular Sciences. 2015; 16(6):14039-14055. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms160614039
Chicago/Turabian StyleLing, Juan, Yanying Zhang, Meilin Wu, Youshao Wang, Junde Dong, Yufeng Jiang, Qingsong Yang, and Siquan Zeng. 2015. "Fungal Community Successions in Rhizosphere Sediment of Seagrasses Enhalus acoroides under PAHs Stress" International Journal of Molecular Sciences 16, no. 6: 14039-14055. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms160614039