Technologies for Single-Cell Isolation
Abstract
:1. Introduction
- Level of automation, distinguishing manual methods for cell separation like microscope-assisted picking from automated devices such as fluorescence activated cell sorters (FACS).
- Ability to isolate specific/individual cells, distinguishing statistical methods (i.e., the separation of cells according to a certain statistical probability) from a controlled cell separation (i.e., a cell is specifically selected and confirmed to be single).
- Compatibility with certain application requirements, distinguishing technologies mainly applied for production of monoclonal cell cultures (derived from single cells) from technologies preferably used for single-cell genome/proteome analysis.
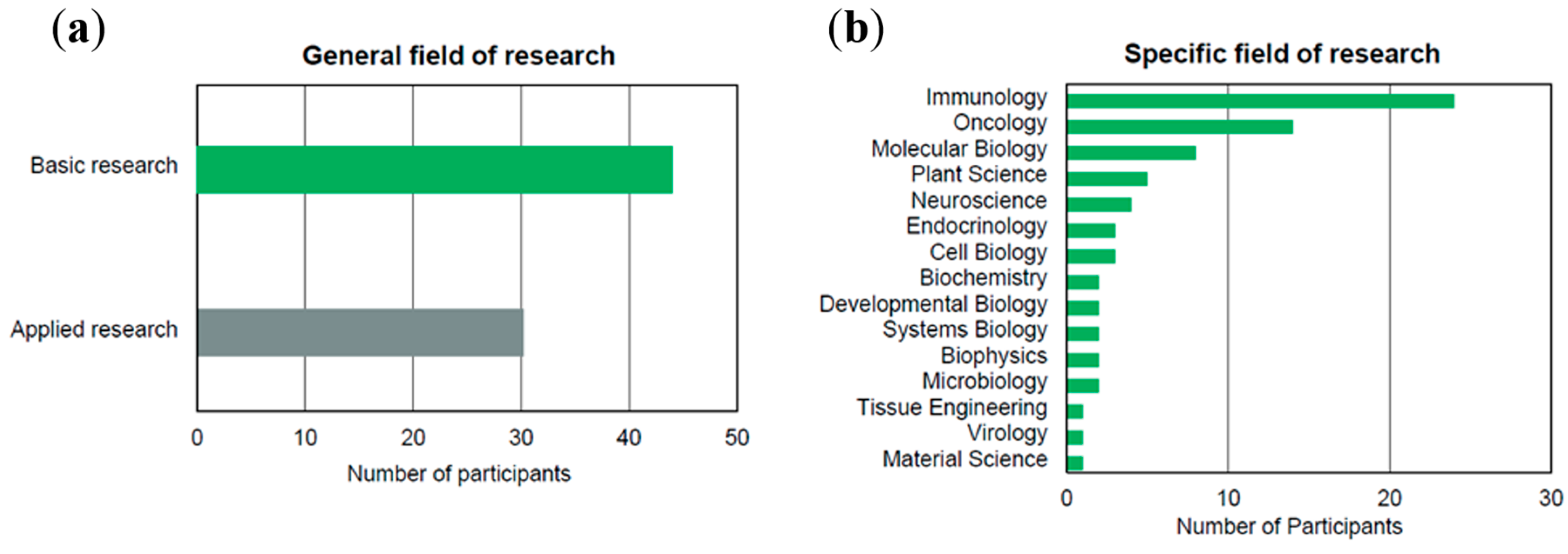
2. Market Study of Single-Cell Technologies

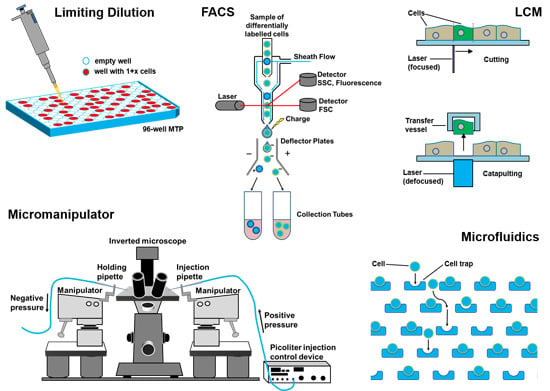
3. Single-Cell Isolation Technologies
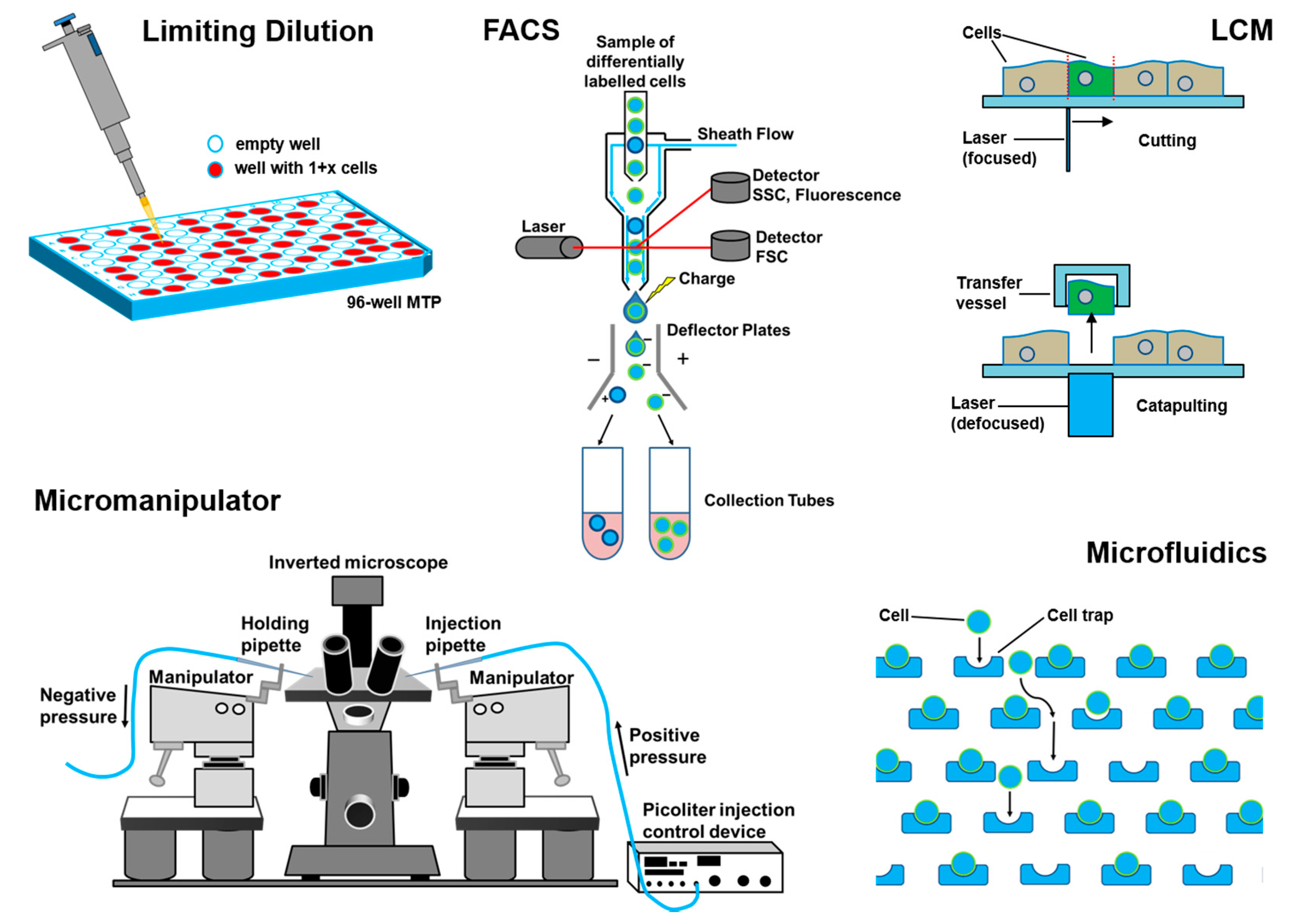
3.1. Flow Cytometry
3.2. Laser Capture Microdissection

3.3. Limiting Dilution
| 0.5 Cells/Aliquot | 0.9 Cells/Aliquot | ||
|---|---|---|---|
| Cell Number/Well | Probability | Cell Number/Well | Probability |
| 0 | 61% | 0 | 41% |
| 1 | 30% | 1 | 37% |
| 2 | 8% | 2 | 16% |
| 3 | 1% | 3 | 5% |
| 4 | 0% | 4 | 1% |
3.4. Manual Cell Picking
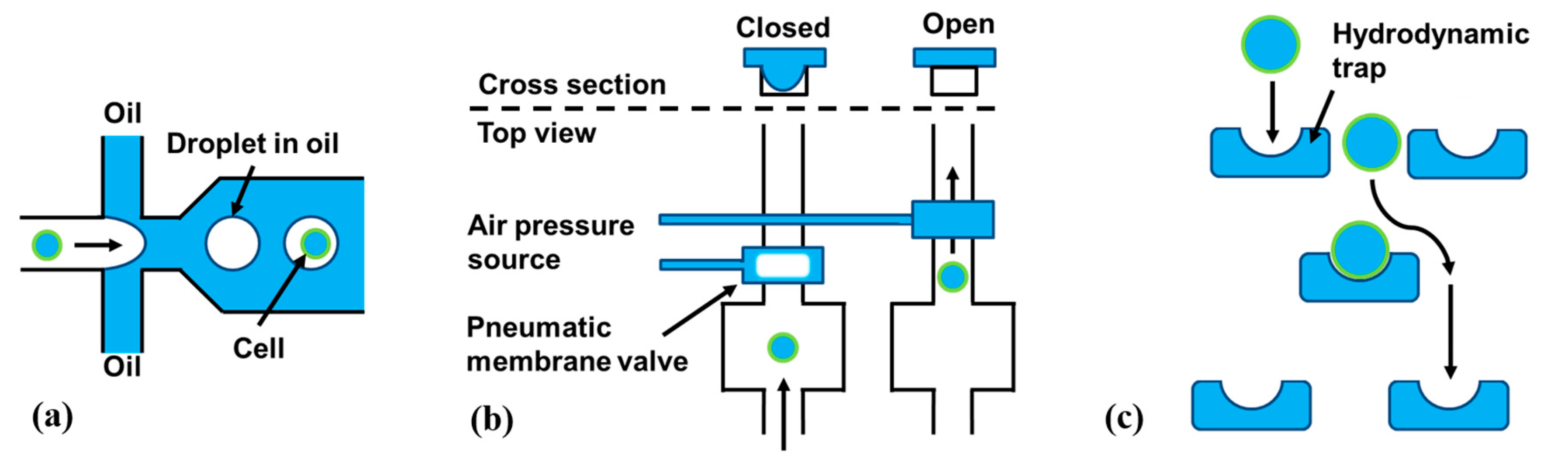
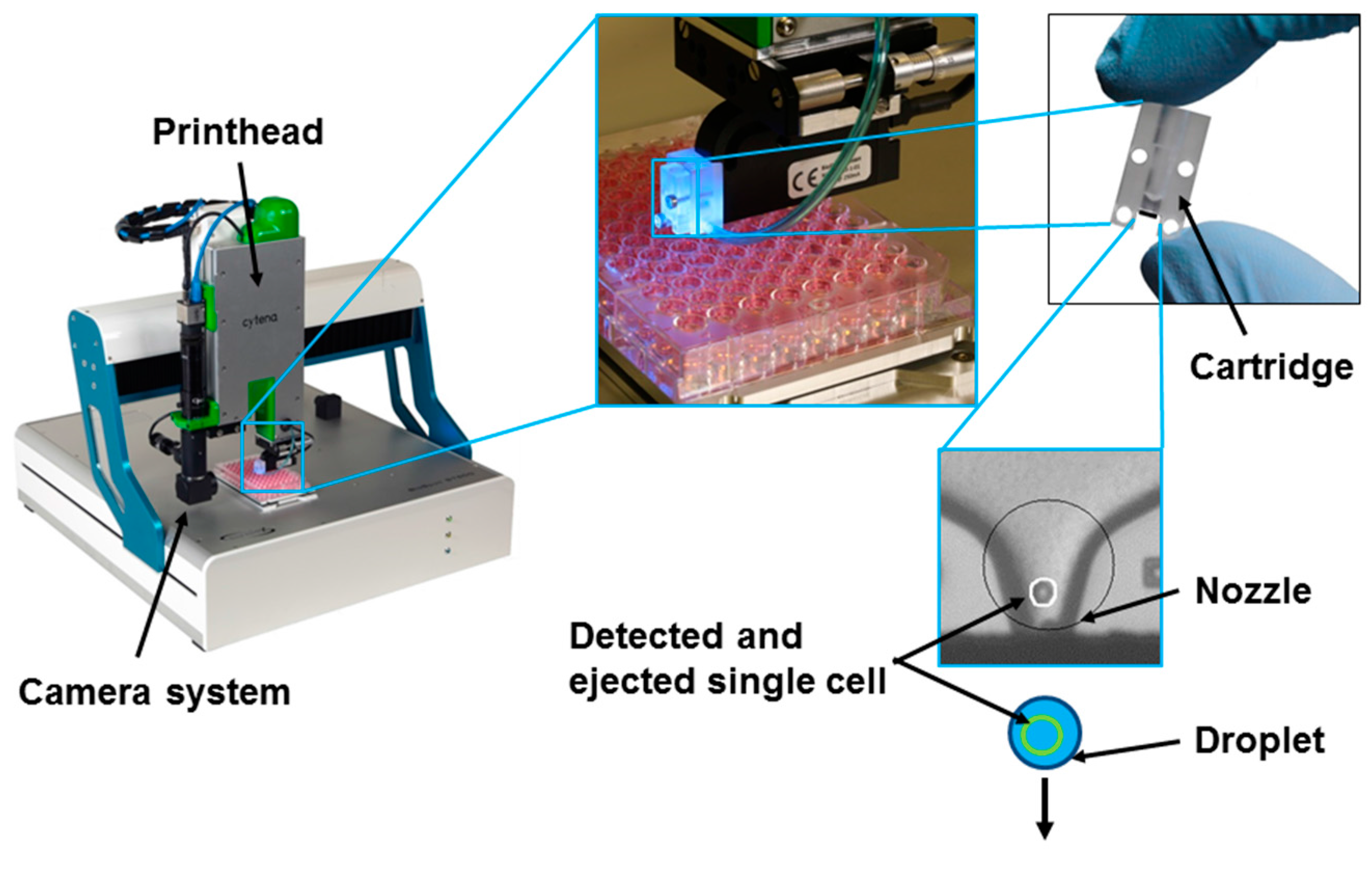
3.5. Microfluidics
4. Patent Search for Single-Cell Separation Technologies
- Only an analysis method of single cells.
- A common method using the term “single cell suspension” without addressing specifically a method for single-cell isolation.
- A cell separation method, which is already established and only part of a patented workflow.
- Other, in this context, irrelevant methods by using the term “cell” in a non-biological context such as for a battery or a chamber in a technical device. Therefore, the terms “fuel” and “solar” were already excluded from the original search from the beginning (see above).
| Title | EPO Publication Number | Reference |
|---|---|---|
| Methods for multiplex analytical measurements in single cells of solid tissues | AU2013315409 (A1) | [55] |
| Single-cell isolation screen adapted with pipettor tip | CN104195036 (A) | [56] |
| An integrated microfluidic device for single-cell isolation, cell lysis and nucleic acid extraction * | CA2817775 (A1) | [57] |
| System and method for capturing and analyzing cells * | US2014349867 (A1) | [58] |
| Single-cell automatic analysis device based on dual-optical-path micro-fluidic chip * | CN203929785 (U) | [59] |
| Microfluidic devices and methods for cell sorting, cell culture and cells based diagnostics and therapeutics * | US2014248621 (A1) | [60] |
| High-throughput single-cell imaging, sorting, and isolation * | US8934700 (B2); US2014247971 (A1) | [61] |
| Automatic single cell analysis method based on microfluidic system * | CN103926190 (A) | [62] |
| Apparatus for single cell separation and position fixing * | US2013129578 (A1); US8475730 (B2) | [63] |
| Method and apparatus for single cell isolation and analysis | US2012315639 (A1) | [64] |
| Apparatus for magnetic separation of cells | US2012045828 (A1) | [65] |
| Method and apparatus for the discretization and manipulation of sample volumes * | CN102187216 (A) | [66] |
| Plate for separating single cell | JP2011152108 (A); JP5622189 (B2) | [67] |
| Array apparatus for separation of single cell * | KR20110037345 (A); KR101252829 (B1) | [68] |
| Device and method for continuously analyzing single-cell contents by miniflow control chip at high speed * | CN101923053 (A); CN101923053 (B) | [69] |
| Complete set of equipment for single cell gel electrophoresis test | CN201662556 (U) | [70] |
| Single cell analysis of membrane molecules * | US2009173631 (A1) | [71] |
| Single-cell inclusion analytical method based on micro-fluidic chip * | CN101393124 (A) | [72] |
| Analytical system based on porous material for highly parallel single cell detection * | US2008020453 (A1) | [73] |
| Single cell isolation apparatus and method of use | US6538810 (B1) | [74] |
| Cell isolation and screening device and method of using same * | WO03011451 (A1) | [75] |
| Cell transfer mechanism and cell fusion apparatus * | JPH0731457 (A) | [76] |
| Device for automatically testing single cell dielectric spectrum based on composite dielectrophoresis | CN201075104 (Y) | [77] |
| High-pass cell separation device and use method therefor * | CN1962845 (A) | [78] |
| Cell inclusions analysis method based on microfluid chip * | CN1734265 (A) | [79] |
5. Future Potential of Single-Cell Technologies

6. Discussion
6.1. General Aspects
6.2. Flow Cytometry
6.3. Laser Capture Microdissection
6.4. Micromanipulator
6.5. Limiting Dilution
6.6. Microfluidics
6.7. Patents and Emerging Technologies
7. Conclusions
| Technology | Automation Level | Throughput | Impact on Cell Integrity | Control over Individual Cell 1 | Compatibility with Established Workflows 2 |
|---|---|---|---|---|---|
| Fluorescence-Activated Cell Sorting (FACS) | Automatic | High | Often impairing | Yes | High |
| Limiting dilution | Manual or automatic | Moderate | Gentle | No | High |
| Micromanipulation | Manual | Low | Moderate | Yes | Moderate |
| Laser-capture microdissection | Manual | Low | Often impairing | Yes | Low |
| Microfluidics (Lab-on-a-Chip) | Automatic | Low to high | Diverse | Typically not | Low |
| Microfluidics (inkjet-like printing) | Automatic | Moderate | Gentle | Yes | High |
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Blainey, P.C.; Quake, S.R. Dissecting genomic diversity, one cell at a time. Nat. Methods. 2014, 11, 19–21. [Google Scholar] [CrossRef] [PubMed]
- Van Loo, P.; Voet, T. Single cell analysis of cancer genomes. Curr. Opin. Genet. Dev. 2014, 24, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Wendl, M.C.; McMichael, J.F.; Raphael, B.J. Expanding the computational toolbox for mining cancer genomes. Nat. Rev. Genet. 2014, 15, 556–570. [Google Scholar] [CrossRef] [PubMed]
- Ilie, M.; Hofman, V.; Long, E.; Bordone, O.; Selva, E.; Washetine, K.; Marquette, C.H.; Hofman, P. Current challenges for detection of circulating tumor cells and cell-free circulating nucleic acids, and their characterization in non-small cell lung carcinoma patients. What is the best blood substrate for personalized medicine? Ann. Transl. Med. 2014, 2, 107. [Google Scholar] [PubMed]
- BIOCOM AG. Biotechnology Database. Available online: http://www.biotechnologie.de/BIO/Navigation/EN/Databases/biotechnology-db.html (accessed on 30 June 2015).
- HTStec. Single Cell Technologies Trends 2014. 2014. Available online: http://selectbiosciences.com/MarketReportsID.aspx?reportID=83 (accessed on 30 June 2015).
- Moldavan, A. Photo-Electric Technique for the Counting of Microscopial Cells; Science: New York, NY, USA, 1934; Volume 80, pp. 188–189. [Google Scholar]
- Gucker, F.T.; O’Konski, C.T. A photoelectronic counter for colloidal particles. J. Am. Chem. Soc. 1947, 69, 2422–2431. [Google Scholar] [CrossRef] [PubMed]
- Coulter, W.H. Means for Counting Particles Suspended in a Fluid. U.S. Patent 2,656,508 A, 20 October 1953. [Google Scholar]
- Fulwyler, M.J. Electronic separation of biological cells by volume. Science 1965, 150, 910–911. [Google Scholar] [CrossRef] [PubMed]
- Fulwyler, M.J. Particle Separator. U.S. Patent 3,380,584 A, 30 April 1968. [Google Scholar]
- Dittrich, W.; Goehde, W. Automatisches Mess- und Zaehlgeraet fuer die Teilchen einer Dispersion. D.E. Patent 1,815,352 A1, 14 January 1971. [Google Scholar]
- Shapiro, H.M. Practical Flow Cytometry,, 4th ed.; Wiley-Liss: New York, NY, USA, 2003. [Google Scholar]
- Kohler, G.; Milstein, C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 1975, 256, 495–497. [Google Scholar] [CrossRef] [PubMed]
- Herzenberg, L.A.; Parks, D.; Sahaf, B.; Perez, O.; Roederer, M.; Herzenberg, L.A. The history and future of the fluorescence activated cell sorter and flow cytometry: A view from Stanford. Clin. Chem. 2002, 48, 1819–1827. [Google Scholar] [PubMed]
- Underwood, A.P.; Bean, P.A. Hazards of the limiting-dilution method of cloning hybridomas. J. Immunol. Methods 1988, 107, 119–128. [Google Scholar] [CrossRef]
- Brown, M.; Wittwer, C. Flow cytometry: Principles and clinical applications in hematology. Clin. Chem. 2000, 46, 1221–1229. [Google Scholar] [PubMed]
- Valet, G. Past and present concepts in flow cytometry: A European perspective. J. Biol. Regul. Homeost. Agents 2003, 17, 213–222. [Google Scholar] [PubMed]
- Davey, H.M.; Kell, D.B. Flow cytometry and cell sorting of heterogeneous microbial populations: The importance of single-cell analyses. Microbiol. Rev. 1996, 60, 641–696. [Google Scholar] [PubMed]
- Lacombe, F.; Belloc, F. Flow cytometry study of cell cycle, apoptosis and drug resistance in acute leukemia. Hematol. Cell Ther. 1996, 38, 495–504. [Google Scholar] [CrossRef] [PubMed]
- McCoy, J.P.; Carey, J.L. Recent advances in flow cytometric techniques for cancer detection and prognosis. Immunol. Ser. 1990, 53, 171–187. [Google Scholar] [PubMed]
- Emmert-Buck, M.R.; Bonner, R.F.; Smith, P.D.; Chuaqui, R.F.; Zhuang, Z.; Goldstein, S.R.; Weiss, R.A.; Liotta, L.A. Laser Capture microdissection. Science 1996, 274, 998–1001. [Google Scholar] [CrossRef] [PubMed]
- Espina, V.; Heiby, M.; Pierobon, M.; Liotta, L.A. Laser capture microdissection technology. Expert Rev. Mol. Diagn. 2007, 7, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Vogel, A.; Noack, J.; Hüttman, G.; Paltauf, G. Mechanisms of femtosecond laser nanosurgery of cells and tissues. Appl. Phys. B 2005, 81, 1015–1047. [Google Scholar] [CrossRef]
- Vogel, A.; Lorenz, K.; Horneffer, V.; Hüttmann, G.; von Smolinski, D.; Gebert, A. Mechanisms of laser-induced dissection and transport of histologic specimens. Biophys. J. 2007, 93, 4481–4500. [Google Scholar] [CrossRef] [PubMed]
- Esposito, G. Complementary techniques: Laser capture microdissection–increasing specificity of gene expression profiling of cancer specimens. Adv. Exp. Med. Boil. 2007, 593, 54–65. [Google Scholar]
- Fink, L.; Kwapiszewska, G.; Wilhelm, J.; Bohle, R.M. Laser-microdissection for cell type- and compartment-specific analyses on genomic and proteomic level. Exp. Toxicol. Pathol. 2006, 5, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Fink, L.; Bohle, R.M. Laser microdissection and RNA analysis. Methods Mol. Biol. 2005, 293, 167–185. [Google Scholar] [PubMed]
- Nakamura, N.; Ruebel, K.; Jin, L.; Qian, X.; Zhang, H.; Lloyd, R.V. Laser capture microdissection for analysis of single cells. Methods Mol. Med. 2007, 132, 11–18. [Google Scholar] [PubMed]
- Keays, K.M.; Owens, G.P.; Ritchie, A.M.; Gilden, D.H.; Burgoon, M.P. Laser capture microdissection and single-cell RT-PCR without RNA purification. J. Immunol. Methods 2005, 302, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Vandewoestyne, M.; Deforce, D. Laser capture microdissection in forensic research: A review. Int. J. Leg. Med. 2010, 124, 513–521. [Google Scholar] [CrossRef] [PubMed]
- DeCarlo, K.; Emley, A.; Dadzie, O.E.; Mahalingam, M. Laser capture microdissection: Methods and applications. Methods Mol. Biol. 2011, 755, 1–15. [Google Scholar] [PubMed]
- Lefkovits, I.; Pernis, B. Immunological Methods; Elsevier Science: Burlington, MA, USA, 1979. [Google Scholar]
- Goding, J.W. Antibody production by hybridomas. J. Immunol. Methods 1980, 285, 285–308. [Google Scholar] [CrossRef]
- Fuller, S.A.; Takahashi, M.; Hurrell, J.G. Cloning of hybridoma cell lines by limiting dilution. Curr. Protoc. Mol. Biol. 2001. [Google Scholar] [CrossRef]
- Yokoyama, W.M.; Christensen, M.; Dos Santos, G.; Miller, D.; Ho, J.; Wu, T.; Dziegelewski, M.; Neethling, F.A. Production of monoclonal antibodies. Curr. Protoc. Immunol. 2013. [Google Scholar] [CrossRef]
- Staszewski, R. Cloning by limiting dilution: An improved estimate that an interesting culture is monoclonal. Yale J. Biol. Med. 1984, 57, 865–868. [Google Scholar] [PubMed]
- Smith, R. Cell technology for cell products. In Proceedings of the 19th ESACT Meeting, Harrogate, UK, 5–8 June 2005.
- Fröhlich, J.; König, H. New techniques for isolation of single prokaryotic cells. FEMS Microbiol. Rev. 2000, 24, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Brehm-stecher, B.F.; Johnson, E.A. Single-cell microbiology: Tools, technologies, and applications. Microbiol. Mol. Biol. Rev. 2004, 68, 538–559. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.; Tucker, M.J.; Morton, P.C.; Sweitzer-Yoder, C.L.; Smith, S.E. Micromanipulation in assisted reproduction: A review of current technology. Curr. Opin. Obstet. Gynecol. 1998, 10, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-X.; Wang, G.-Q.; Li, W.-S.; Huang, J.-P.; Ji, A.-Q.; Hu, L. New cell separation technique for the isolation and analysis of cells from biological mixtures in forensic caseworks. Croat. Med. J. 2011, 52, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Sims, C.E.; Allbritton, N.L. Analysis of single mammalian cells on-chip. Lab Chip 2007, 7, 423–440. [Google Scholar] [CrossRef] [PubMed]
- Lecault, V.; White, A.K.; Singhal, A.; Hansen, C.L. Microfluidic single cell analysis: From promise to practice. Curr. Opin. Chem. Biol. 2012, 16, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Gossett, D.R.; Weaver, W.M.; Mach, A.J.; Hur, S.C.; Tse, H.T.K.; Lee, W.; Amini, H.; di Carlo, D. Label-free cell separation and sorting in microfluidic systems. Anal. Bioanal. Chem. 2010, 397, 3249–3267. [Google Scholar] [CrossRef] [PubMed]
- Brouzes, E.; Medkova, M.; Savenelli, N.; Marran, D.; Twardowski, M.; Hutchison, J.B.; Rothberg, J.M.; Link, D.R.; Perrimon, N.; Samuels, M.L. Droplet microfluidic technology for single-cell high-throughput screening. Proc. Natl. Acad. Sci. USA 2009, 106, 14195–14200. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Sjoberg, R.; Leyrat, A.A.; Pirone, D.M.; Chen, C.S.; Quake, S.R. Versatile, fully automated, microfluidic cell culture system. Anal.Chem. 2007, 79, 8557–8563. [Google Scholar] [CrossRef] [PubMed]
- Di Carlo, D.; Wu, L.Y.; Lee, L.P. Dynamic single cell culture array. Lab Chip 2006, 6, 1445–1449. [Google Scholar] [CrossRef] [PubMed]
- Edd, J.F.; di Carlo, D.; Humphry, K.J.; KÇôster, S.; Irimia, D.; Weitz, D.; Toner, M. Controlled encapsulation of single-cells into monodisperse picolitre drops. Lab Chip 2008, 8, 1262–1264. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Han, X.; Li, Y.; Li, S.Y.; Zu, Y.; Wang, Z.; Qin, L. Hand-held and integrated single-cell pipettes. J. Am. Chem. Soc. 2014, 136, 10858–10861. [Google Scholar] [CrossRef] [PubMed]
- Ateya, D.A.; Erickson, J.S.; Howell, P.B.; Hilliard, L.R.; Golden, J.P.; Ligler, F.S. The good, the bad, and the tiny: A review of microflow cytometry. Anal. Bioanal. Chem 2008, 391, 1485–1498. [Google Scholar] [CrossRef] [PubMed]
- Yusof, A.; Keegan, H.; Spillane, C.D.; Sheils, O.M.; Martin, C.M.; O’Leary, J.J.; Zengerle, R.; Koltay, P. Inkjet-like printing of single-cells. Lab Chip 2011, 11, 2447–2454. [Google Scholar] [CrossRef] [PubMed]
- Gross, A.; Schondube, J.; Niekrawitz, S.; Streule, W.; Riegger, L.; Zengerle, R.; Koltay, P. Single-cell printer: Automated, on demand, and label free. J. Lab. Autom. 2013, 18, 504–518. [Google Scholar] [CrossRef] [PubMed]
- Stumpf, F.; Schoendube, J.; Gross, A.; Rath, C.; Niekrawietz, S.; Koltay, P.; Roth, G. Single-cell PCR of genomic DNA enabled by automated single-cell printing for cell isolation. Biosens. Bioelectron. 2015, 69, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Finski, A.; Macbeath, G. Methods for Multiplex Analytical Measurements in Single Cells of Solid Tissues. Patent AU2013315409 (A1), 2 April 2015. [Google Scholar]
- Liu, D. Single-Cell Isolation Screen Adapted with Pipettor Tip. Patent CN104195036 (A), 10 December 2014. [Google Scholar]
- Yao, C.D. An Integrated Microfluidic Device for Single-Cell Isolation, Cell Lysis and Nucleic Acid Extraction. Patent CA2817775 (A1), 29 November 2014. [Google Scholar]
- Handique, K.; Gogoi, P.; Javdani, S.S.; Zhou, Y. System and Method for Capturing and Analyzing Cells. US2014349867 (A1), 27 November 2014. [Google Scholar]
- Liu, Y.; Li, J.; Chen, Y.; Jian, D.; Gao, J. Single-Cell Automatic Analysis Device Based on Dual-Optical-Path Micro-Fluidic Chip . Patent CN203929785 (U), 5 November 2014. [Google Scholar]
- Collins, J. Microfluidic Devices and Methods for Cell Sorting, Cell Culture and Cells Based Diagnostics and Therapeutics. Patent US2014248621 (A1), 4 September 2014. [Google Scholar]
- Bharadwaj, R.; Fathollahi, B. High-Throughput Single-Cell Imaging, Sorting, and Isolation. Patent US8934700 (B2), US2014247971 (A1), 4 September 2014. [Google Scholar]
- Liu, Y.; Ban Qing, G.J. Automatic Single Cell Analysis Method Based on Microfluidic System. Patent CN103926190 (A), 16 July 2014. [Google Scholar]
- Jeong, O.C. Apparatus for Single Cell Separation and Position Fixing. Patent US2013129578 (A1), US8475730 (B2), 23 May 2013. [Google Scholar]
- Deng, G.Y.; Zhang, J.; Tian, F. Method and Apparatus for Single Cell Isolation and Analysis. Patent US2012315639 (A1), 13 December 2012. [Google Scholar]
- Davis, R.W.; Jeffrey, S.S.; Mindrinos, M.N.; Pease, R.F.; Powell, A.A.; Talasaz, A.H. Apparatus for Magnetic Separation of Cells. Patent US2012045828 (A1), 23 February 2012. [Google Scholar]
- Chiu, D.T.; Cohen, D.E.; Jeffries, G.D.M. Method and Apparatus for the Discretization and Manipulation of Sample Volumes. Patent CN102187216 (A), 14 September 2011. [Google Scholar]
- Matsutani, A.; Takada, A. Plate for Separating Single Cell. Patent JP2011152108 (A), JP5622189 (B2), 11 August 2011. [Google Scholar]
- Park, J.W.; Jung, M.Y.; Park, S.H. Array Apparatus for Separation of Single Cell. Patent KR20110037345 (A), KR101252829 (B1), 13 April 2011. [Google Scholar]
- Yin, X.F.; Xu, C.X.; Liu, J.H. Device and Method for Continuously Analyzing Single-Cell Contents by Miniflow Control Chip at High Speed. Patent CN101923053 (A), CN101923053 (B), 22 December 2010. [Google Scholar]
- Zhang, C.K.; Shi, J.Y.; Zeng, Y.C.; Shen, C.F.; Chen, Y.X.; Chen, L. Complete Set of Equipment for Single Cell Gel Electrophoresis Test. Patent CN201662556 (U), 1 December 2010. [Google Scholar]
- Boone, T.; Singh, S. Single Cell Analysis of Membrane Molecules. Patent US2009173631 (A1), 9 July 2009. [Google Scholar]
- Wu, D.P.; Yu, L.F.; Lin, B.C.; Qin, J.H. Single-Cell Inclusion Analytical Method Based on Micro-Fluidic Chip . Patent CN101393124 (A), 25 March 2009. [Google Scholar]
- Ehben, T.; Zilch, C. Analytical System Based on Porous Material for Highly Parallel Single Cell Detection. Patent US2008020453 (A1), 24 January 2008. [Google Scholar]
- Karanfilov, C. Single Cell Isolation Apparatus and Method of Use. Patent US6538810 (B1), 25 March 2003. [Google Scholar]
- Wang, E.; Kim, E.; Campbell, S.; Kirk, G.L.; Casagrande, R. Cell Isolation and Screening Device and Method of Using Same. Patent WO03011451 (A1), 13 February 2003. [Google Scholar]
- Suzuki, H.; Yasunaka, T. Cell Transfer Mechanism and Cell Fusion Apparatus. Patent JPH0731457 (A), 3 February 1995. [Google Scholar]
- Ni, Z.H.; Song, C.F.; Yi, H.; Zhu, S.C. Device for Automatically Testing Single Cell Dielectric Spectrum Based on Composite Dielectrophoresis. Patent CN201075104 (Y), 18 June 2008. [Google Scholar]
- Shen, B.Y. High-Pass Cell Separation Device and Use Method Therefor. Patent CN1962845 (A), 16 May 2007. [Google Scholar]
- Lin, B.Y. Cell Inclusions Analysis Method Based on Microfluid Chip. Patent CN1734265 (A), 15 February 2006. [Google Scholar]
- De Souza, N. Single-cell methods. Nat. Methods 2011, 9, 35. [Google Scholar] [CrossRef]
- Editorial. Method of the Year 2013. Nat. Methods 2013, 11, 1. [Google Scholar]
- Shapiro, E.; Biezuner, T.; Linnarsson, S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat. Rev. Genet. 2013, 14, 618–630. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Stott, S.; Toner, M.; Maheswaran, S.; Haber, D.A. Circulating tumor cells: Approaches to isolation and characterization. J. Cell Biol. 2011, 192, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Singh, A.K. Single-cell protein analysis. Curr. Opin. Biotechnol. 2012, 23, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Mardis, E.R. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008, 24, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Jahan-Tigh, R.R.; Ryan, C.; Obermoser, G.; Schwarzenberger, K. Flow cytometry. J. Investig. Dermatol. 2012, 132. [Google Scholar] [CrossRef] [PubMed]
- Mollet, M.; Godoy-Silva, R.; Berdugo, C.; Chalmers, J.J. Computer simulations of the energy dissipation rate in a fluorescence-activated cell sorter: Implications to cells. Biotechnol. Bioeng. 2008, 100, 260–272. [Google Scholar] [CrossRef] [PubMed]
- Bevilacqua, C.; Makhzami, S.; Helbling, J.-C.; Defrenaix, P.; Martin, P. Maintaining RNA integrity in a homogeneous population of mammary epithelial cells isolated by Laser Capture Microdissection. BMC Cell Biol. 2010, 11, 95. [Google Scholar] [CrossRef] [PubMed]
- Liu, A. Laser capture microdissection in the tissue biorepository. J. Biomol. Tech. 2010, 21, 120–125. [Google Scholar] [PubMed]
- Fend, F. Laser capture microdissection in pathology. J. Clin. Pathol. 2000, 53, 666–672. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Moraes, C.; Zhao, Y.; You, L.D.; Simmons, C.A.; Sun, Y. A micromanipulation system for single cell deposition. In Proceeding of 2010 IEEE International Conference on Robotics and Automation (ICRA 2010), Anchorage, Alaska, 3–8 May 2010; IEEE: New York, NY, USA, May 2010; pp. 494–499. [Google Scholar]
- Blow, N. Microfluidics: In search of a killer application. Nat. Methods 2007, 4, 665–668. [Google Scholar] [CrossRef]
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Becker, H. Hype, hope and hubris: The quest for the killer application in microfluidics. Lab Chip 2009, 9, 2119–2122. [Google Scholar] [CrossRef] [PubMed]
- Haber, C. Microfluidics in commercial applications; an industry perspective. Lab Chip 2006, 6, 1118–1121. [Google Scholar] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gross, A.; Schoendube, J.; Zimmermann, S.; Steeb, M.; Zengerle, R.; Koltay, P. Technologies for Single-Cell Isolation. Int. J. Mol. Sci. 2015, 16, 16897-16919. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms160816897
Gross A, Schoendube J, Zimmermann S, Steeb M, Zengerle R, Koltay P. Technologies for Single-Cell Isolation. International Journal of Molecular Sciences. 2015; 16(8):16897-16919. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms160816897
Chicago/Turabian StyleGross, Andre, Jonas Schoendube, Stefan Zimmermann, Maximilian Steeb, Roland Zengerle, and Peter Koltay. 2015. "Technologies for Single-Cell Isolation" International Journal of Molecular Sciences 16, no. 8: 16897-16919. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms160816897