Metabolite Profiling of Diverse Rice Germplasm and Identification of Conserved Metabolic Markers of Rice Roots in Response to Long-Term Mild Salinity Stress
Abstract
:1. Introduction
2. Results
2.1. Growth Potential of 38 Rice Genotypes under Long-Term Mild Salinity Stress

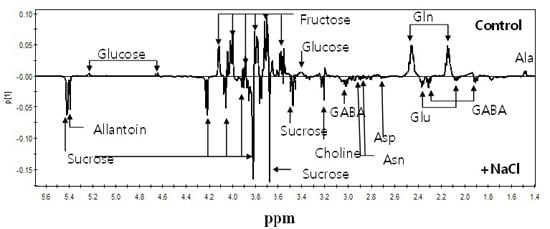
2.2. 1H-NMR Spectroscopy of Root Metabolites of 38 Rice Genotypes and Multivariate Statistical Analyses Relating to the Growth Rate and Salt Response of Rice Roots


2.3. Metabolic Profiling of 38 Rice Genotypes Revealed Conserved Salt Stress Metabolite Markers of Rice Roots


3. Discussion
3.1. The Glutamate/Glutamine Pathway Plays an Important Role in Rice Roots Exposed to Long-Term Mild Salt Stress
3.2. Sucrose Accumulation Is a Conserved Metabolic Response in Rice Roots Exposed to Long-Term Mild Salinity
3.3. Allantoin Is a Potential Metabolic Marker of Rice Tolerant to Growth Stress
4. Experimental Section
4.1. Rice Growth and Salt Stress Treatment
4.2. Metabolite Extraction from Rice Roots
4.3. NMR Measurements
4.4. Data Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
References
- Akbar, M.; Yabuno, T.; Nakao, S. Breeding for saline-resistant varieties of rice. I. Variability for salt tolerance among some rice varieties. Jpn. J. Breed. 1972, 22, 277–284. [Google Scholar] [CrossRef]
- Flowers, T.J.; Yeo, A.R. Variability in the resistance of sodium chloride salinity within rice (Oryza sativa L.) varieties. New Phytol. 1981, 88, 363–373. [Google Scholar] [CrossRef]
- Kahn, M.S.A.; Hamid, A.; Karim, M.A. Effect of sodium chloride on germination and seedling characters of different types of rice (Oryza sativa L.). J. Agron. Crop Sci. 1997, 179, 163–169. [Google Scholar] [CrossRef]
- Lutts, S.; Kinet, J.M.; Bouharmout, J. Changes in plant response to NaCl during development of rice (Oryza sativa L.) varieties differing in salinity resistance. J. Exp. Bot. 1995, 46, 1843–1852. [Google Scholar] [CrossRef]
- Yoshida, S. Fundamentals of Rice Crop Science; IRRI: Los Banos, Philippines, 1981; pp. 175–176. [Google Scholar]
- Omokawa, H.; Aonuma, S. Amelioration of the salt-stressed root growth of rice and normalization of the Na+ distribution between the shoot and root by (S)-α-methylbenzyl-2-fluoro-4-methylphenylurea. Biosci. Biotechnol. Biochem. 2002, 66, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Ji, H.; Pardo, J.M.; Batelli, G.; van Oosten, M.J.; Bressan, R.A.; Li, X. The salt overly sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Atienza, J.; Jiang, X.; Garciadeblas, B.; Mendoza, I.; Zhu, J.K.; Pardo, J.M.; Quintero, F.J. Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 2007, 143, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, K.; Hwang, H.; Bhatnagar, N.; Kim, D.Y.; Yoon, I.S.; Byun, M.O.; Kim, S.T.; Jung, K.H.; Kim, B.G. Overexpression of PYL5 in rice enhances drought tolerance, inhibits growth, and modulates gene expression. J. Exp. Bot. 2014, 65, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Mao, B.; Ou, S.; Wang, W.; Liu, L.; Wu, Y.; Chu, C.; Wang, X. OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol. Biol. 2014, 84, 19–36. [Google Scholar] [CrossRef] [PubMed]
- Diédhiou, C.J.; Popova, O.V.; Dietz, K.J.; Golldack, D. The SNF1-type serine-threonine protein kinase SAPK4 regulates stress-responsive gene expression in rice. BMC Plant Biol. 2008, 28, 49. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O. Metabolomics: The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Bamba, T.; Harada, K.; Fukusaki, E.; Kobayashi, A. Time-course metabolic profiling in Arabidopsis thaliana cell cultures after salt stress treatment. J. Exp. Bot. 2007, 58, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Choi, Y.H.; Verpoorte, R. NMR-based metabolomic analysis of plants. Nat. Protoc. 2010, 5, 536–549. [Google Scholar] [CrossRef] [PubMed]
- Ali, K.; Iqbal, M.; Fortes, A.M.; Pais, M.S.; Korthout, H.A.; Verpoorte, R.; Choi, Y.H. Red wines attenuate TNFα production in human histiocytic lymphoma cell line: An NMR spectroscopy and chemometrics based study. Food Chem. 2013, 141, 3124–3130. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, Z.; Yang, S.; Wang, Y.; Yu, L.; Zhang, B.; Rao, Z.; Gao, J.; Tu, S. 1H-NMR-based metabolomic analysis for identifying serum biomarkers to evaluate methotrexate treatment in patients with early rheumatoid arthritis. Exp. Ther. Med. 2012, 4, 165–171. [Google Scholar] [PubMed]
- Teixeira, J.; Fidalgo, F. Salt stress affects glutamine synthetase activity and mRNA accumulation on potato plants in an organ-dependent manner. Plant Physiol. Biochem. 2009, 47, 807–813. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, M.; Guo, R.; Shi, D.; Liu, B.; Lin, X.; Yang, C. Effects of salt stress on ion balance and nitrogen metabolism of old and young leaves in rice (Oryza sativa L.). BMC Plant Biol. 2012, 12, 194. [Google Scholar] [CrossRef] [PubMed]
- Funayama, K.; Kojima, S.; Tabuchi-Kobayashi, M.; Sawa, Y.; Nakayama, Y.; Hayakawa, T.; Yamaya, T. Cytosolic glutamine synthetase 1;2 is responsible for the primary assimilation of ammonium in rice roots. Plant Cell Physiol. 2013, 54, 934–943. [Google Scholar] [CrossRef] [PubMed]
- Kusano, M.; Tabuchi, M.; Fukushima, A.; Funayama, K.; Diaz, C.; Kobayashi, M.; Hayashi, N.; Tsuchiya, Y.N.; Takahashi, H.; Kamata, A.; et al. Metabolomics data reveal a crucial role of cytosolic glutamine synthetase 1;1 in coordinating metabolic balance in rice. Plant J. 2011, 66, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Siahpoosh, M.R.; Sanchez, D.H.; Schlereth, A.; Scofield, G.N.; Furbank, R.T.; van Dongen, J.T.; Kopka, J. Modification of OsSUT1 gene expression modulates the salt response of rice Oryza sativa cv. Taipei 309. Plant Sci. 2012, 182, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Zhou, Y.; Xiao, J.; Li, X.; Zhang, Q.; Lian, X. Overexpressed glutamine synthetase gene modifies nitrogen metabolism and abiotic stress responses in rice. Plant Cell Rep. 2009, 28, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T.; Satoh, Y.; Ohki, A.; Takada, R.; Arai, T.; Michiyama, H. Inhibition of ammonium assimilation restores elongation of seminal rice roots repressed by high levels of exogenous ammonium. Physiol. Plant. 2008, 134, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Rosa, M.; Prado, C.; Podazza, G.; Interdonato, R.; González, J.A.; Hilal, M.; Prado, F.E. Soluble sugars—Metabolism, sensing and abiotic stress: A complex network in the life of plants. Plant Signal. Behav. 2009, 4, 388–393. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, D.H.; Siahpoosh, M.R.; Roessner, U.; Udvardi, M.; Kopka, J. Plant metabolomics reveals conserved and divergent metabolic responses to salinity. Physiol. Plant. 2008, 132, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Ford, K.L.; Roessner, U.; Natera, S.; Cassin, A.M.; Patterson, J.H.; Bacic, A. Rice suspension cultured cells are evaluated as a model system to study salt responsive networks in plants using a combined proteomic and metabolomic profiling approach. Proteomics 2013, 13, 2046–2062. [Google Scholar] [CrossRef] [PubMed]
- Lemoine, R.; la Camera, S.; Atanassova, R.; Dédaldéchamp, F.; Allario, T.; Pourtau, N.; Bonnemain, J.L.; Laloi, M.; Coutos-Thévenot, P.; Maurousset, L.; et al. Source-to-sink transport of sugar and regulation by environmental factors. Front. Plant Sci. 2013, 4, 272. [Google Scholar] [CrossRef] [PubMed]
- Kanemura, T.; Homma, K.; Ohsumi, A.; Shiraiwa, T.; Horie, T. Evaluation of genotypic variation in leaf photosynthetic rate and its associated factors by using rice diversity research set of germplasm. Photosynth. Res. 2007, 94, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Boriboonkaset, T.; Theerawitaya, C.; Yamada, N.; Pichakum, A.; Supaibulwatana, K.; Cha-Um, S.; Takabe, T.; Kirdmanee, C. Regulation of some carbohydrate metabolism-related genes, starch and soluble sugar contents, photosynthetic activities and yield attributes of two contrasting rice genotypes subjected to salt stress. Protoplasma 2013, 250, 1157–1167. [Google Scholar] [CrossRef] [PubMed]
- Chung, W.Y.; Benzie, I.F. Plasma allantoin measurement by isocratic liquid chromatography with tandem mass spectrometry: Method evaluation and application in oxidative stress biomonitoring. Clin. Chim. Acta 2013, 424, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Werner, A.K.; Witte, C.P. The biochemistry of nitrogen mobilization: Purine ring catabolism. Trends Plant Sci. 2011, 16, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Tegeder, M. Transporters involved in source to sink partitioning of amino acids and ureides: Opportunities for crop improvement. J. Exp. Bot. 2014, 65, 1865–1878. [Google Scholar] [CrossRef] [PubMed]
- Collier, R.; Tegeder, M. Soybean ureide transporters play a critical role in nodule development, function and nitrogen export. Plant J. 2012, 72, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Brychkova, G.; Alikulov, Z.; Fluhr, R.; Sagi, M. A critical role for ureides in dark and senescence-induced purine remobilization is unmasked in the Atxdh1 Arabidopsis mutant. Plant J. 2008, 54, 496–509. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Kong, C.H.; Sun, B.; Xu, X.H. Distribution and function of allantoin (5-ureidohydantoin) in rice grains. J. Agric. Food Chem. 2012, 60, 2793–2798. [Google Scholar] [CrossRef] [PubMed]
- Rose, M.T.; Rose, T.J.; Pariasca-Tanaka, J.; Yoshihashi, T.; Neuweger, H.; Goesmann, A.; Frei, M.; Wissuwa, M. Root metabolic response of rice (Oryza sativa L.) genotypes with contrasting tolerance to zinc deficiency and bicarbonate excess. Planta 2012, 236, 959–973. [Google Scholar] [CrossRef] [PubMed]
- Degenkolbe, T.; Do, P.T.; Kopka, J.; Zuther, E.; Hincha, D.K.; Köhl, K.I. Identification of drought tolerance markers in a diverse population of rice cultivars by expression and metabolite profiling. PLoS ONE 2013, 8, e63637. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S.; Forno, D.; Cook, J.; Gomez, K. Laboratory Manual for Physiological Studies of Rice; International Rice Research Institute: Laguna, Philippines, 1976; pp. 61–66. [Google Scholar]
- Choi, H.K.; Choi, Y.H.; Verberne, M.; Lefeber, A.W.; Erkelens, C.; Verpoorte, R. Metabolic fingerprinting of wild type and transgenic tobacco plants by 1H-NMR and multivariate analysis technique. Phytochemistry 2004, 65, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Baker, J.M.; Hawkins, N.D.; Ward, J.L.; Lovegrove, A.; Napier, J.A.; Shewry, P.R.; Beale, M.H. A metabolomic study of substantial equivalence of field-grown genetically modified wheat. Plant Biotechnol. J. 2006, 4, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.M.T. Metabolite profiling by one- and two-dimensional NMR analysis of complex mixtures. Prog. Nucl. Magn. Reson. Spectrosc. 1996, 28, 161–219. [Google Scholar] [CrossRef]
- Larsen, F.H.; van den Berg, F.; Engelsen, S.B. An exploratory chemometric study of 1H-NMR spectra of table wine. J. Chemom. 2006, 20, 198–208. [Google Scholar] [CrossRef]
- Eriksson, L.; Johansson, E.; Kettaneh-Wold, N.; Trygg, J.; Wikstrom, C.; Wold, S. Multi and Megavariate Data Analysis Part I: Basic Principles and Applications, 2nd ed.; Umetrics: Umea, Sweden, 2006. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nam, M.H.; Bang, E.; Kwon, T.Y.; Kim, Y.; Kim, E.H.; Cho, K.; Park, W.J.; Kim, B.-G.; Yoon, I.S. Metabolite Profiling of Diverse Rice Germplasm and Identification of Conserved Metabolic Markers of Rice Roots in Response to Long-Term Mild Salinity Stress. Int. J. Mol. Sci. 2015, 16, 21959-21974. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms160921959
Nam MH, Bang E, Kwon TY, Kim Y, Kim EH, Cho K, Park WJ, Kim B-G, Yoon IS. Metabolite Profiling of Diverse Rice Germplasm and Identification of Conserved Metabolic Markers of Rice Roots in Response to Long-Term Mild Salinity Stress. International Journal of Molecular Sciences. 2015; 16(9):21959-21974. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms160921959
Chicago/Turabian StyleNam, Myung Hee, Eunjung Bang, Taek Yun Kwon, Yuran Kim, Eun Hee Kim, Kyungwon Cho, Woong June Park, Beom-Gi Kim, and In Sun Yoon. 2015. "Metabolite Profiling of Diverse Rice Germplasm and Identification of Conserved Metabolic Markers of Rice Roots in Response to Long-Term Mild Salinity Stress" International Journal of Molecular Sciences 16, no. 9: 21959-21974. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms160921959