Alternative Splicing in Adhesion- and Motility-Related Genes in Breast Cancer
Abstract
:1. Introduction
2. Results
2.1. MCF-7 Transcriptome and Alternative Splicing Profile

| Gene Name | Positions | ID | Skipped Exons | ORF Change | Affected Domain(s) | Protein Length |
|---|---|---|---|---|---|---|
| THBS1 | 15q14 | LN607833 | 10 | Del | TSP1 | 1112 |
| LGALS1 | 22q13.1 | LN607840 | 3 | Frameshift | β-galactoside binding | 39 |
| RAP1GAP | 1p36.12 | LN607839 | 24 | Ins | N/A | 821 |
| CD47 | 3q13.12 | LN680437 | 8–10 | Frameshift | Cytoplasmic C-term | 293 |
| RHOA | 3p21.31 | LN607834 | 3–4 | Del | GTP binding | 109 |
| RHOD | 11q13.2 | LN607835 | 4 | Del | GTP binding | 165 |
| CASK | Xp11.4 | LN607837 | 19–20 | Del | Linker between PDZ and SH3 | 886 |
| CTTN | 11q13.3 | LN607836 | 10–11 | Del | Cortactin | 476 |
| JAG2 | 14q32.33 | LN607838 | 24 | Del | VWFC | 1194 |
| SEMA3C | 7q21.11 | LN626689 | 15 | Frameshift | Type 1 Ig-like and R/K rich | 514 |
| SEMA3F | 3p21.31 | LN626688 | 16 | Frameshift | Type 1 Ig-like and R/K rich | 571 |
| PLXNB1 | 3p21.31 | LN626690 | 32 | Frameshift | Cytoplasmic C-term | 1876 |
2.2. New Transcripts of Adhesion-Related Proteins

2.3. New Transcripts of Motility-Related Proteins

2.4. New Semaphorins and Plexins Isoforms

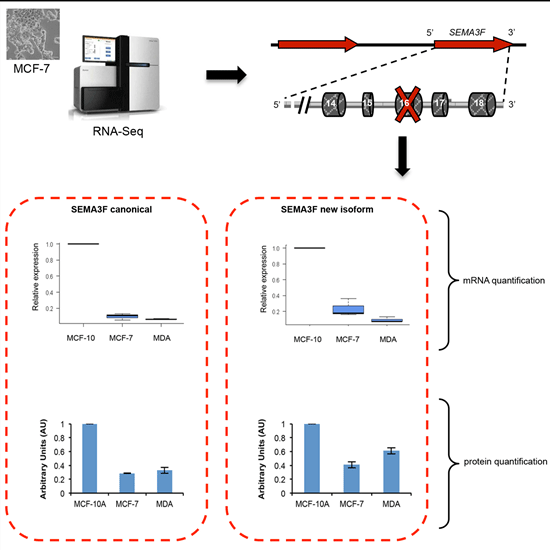
2.5. A New Semaphorin 3F Isoform Expressed in Breast Cancer


3. Discussion
4. Experimental Section
4.1. Cell Lines and Tissue Samples
4.2. RNA Isolation and RNA-Sequencing
4.3. In Silico Analysis
4.4. RT-PCR, Cloning and Sequencing
4.5. qRT PCR
4.6. Western Blot and Immunohistochemistry
4.7. Statistics
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Ervik, M.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. GLOBOCAN 2012: Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012. Available online: http://globocan.iarc.fr (accessed on 29 November 2014).
- Assinder, S.J.; Au, E.; Dong, Q.; Winnick, C. A novel splice variant of the β-tropomyosin (TPM2) gene in prostate cancer. Mol. Carcinog. 2010, 49, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Valastyan, S.; Weinberg, R.A. Tumor metastasis: Molecular insights and evolving paradigms. Cell 2011, 147, 275–292. [Google Scholar] [CrossRef] [PubMed]
- Palmer, T.D.; Ashby, W.J.; Lewis, J.D.; Zijlstra, A. Targeting tumor cell motility to prevent metastasis. Adv. Drug Deliv. Rev. 2011, 63, 568–581. [Google Scholar] [CrossRef] [PubMed]
- D’Apice, L.; Costa, V.; Valente, C.; Trovato, M.; Pagani, A.; Manera, S.; Regolo, L.; Zambelli, A.; Ciccodicola, A.; de Berardinis, P. Analysis of SEMA6B gene expression in breast cancer: Identification of a new isoform. Biochim. Biophys. Acta 2013, 1830, 4543–4553. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Gupta, R.; Davuluri, R.V. Alternative transcription and alternative splicing in cancer. Pharmacol. Ther. 2012, 136, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Eswaran, J.; Horvath, A.; Godbole, S.; Reddy, S.D.; Mudvari, P.; Ohshiro, K.; Cyanam, D.; Nair, S.; Fuqua, S.A.; Polyak, K.; et al. RNA sequencing of cancer reveals novel splicing alterations. Sci. Rep. 2013, 3, 1689. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lee, W.; Jiang, Z.; Chen, Z.; Jhunjhunwala, S.; Haverty, P.M.; Gnad, F.; Guan, Y.; Gilbert, H.N.; Stinson, J.; et al. Genome and transcriptome sequencing of lung cancers reveal diverse mutational and splicing events. Genome Res. 2012, 22, 2315–2327. [Google Scholar] [CrossRef] [PubMed]
- Scarpato, M.; Esposito, R.; Evangelista, D.; Aprile, M.; Ambrosio, M.R.; Angelini, C.; Ciccodicola, A.; Costa, V. AnaLysis of Expression on human chromosome 21, ALE-HSA21: A pilot integrated web resource. Database (Oxford) 2014. [Google Scholar] [CrossRef] [PubMed]
- Chekhun, S.; Bezdenezhnykh, N.; Shvets, J.; Lukianova, N. Expression of biomarkers related to cell adhesion, metastasis and invasion of breast cancer cell lines of different molecular subtype. Exp. Oncol. 2013, 35, 174–179. [Google Scholar] [PubMed]
- Worzfeld, T.; Swiercz, J.; Looso, M.; Straub, B.K.; Sivaraj, K.K.; Offermanns, S. ErbB-2 signals through Plexin-B1 to promote breast cancer metastasis. J. Clin. Investig. 2012, 122, 1296–1305. [Google Scholar] [CrossRef] [PubMed]
- Miyato, H.; Tsuno, N.H.; Kitayama, J. Semaphorin 3C is involved in the progression of gastric cancer. Cancer Sci. 2012, 103, 1961–1966. [Google Scholar] [CrossRef] [PubMed]
- Mendes-da-Cruz, D.A.; Brignier, A.C.; Asnafi, V.; Baleydier, F.; Messias, C.V.; Lepelletier, Y.; Bedjaoui, N.; Renand, A.; Smaniotto, S.; Canioni, D.; et al. Semaphorin SEMA3F and Neuropilin-2 control the migration of human T-cell precursors. PLoS ONE 2014, 9, e103405. [Google Scholar] [CrossRef] [PubMed]
- Goldblum, S.E.; Young, B.A.; Wang, P.; Murphy-Ullrich, J.E. Thrombospondin-1 induces tyrosine phosphorylation of adherens junction proteins and regulates an endothelial paracellular pathway. Mol. Biol. Cell 1999, 10, 1537–1551. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Gao, L.; Feng, Y.; Yuan, L.; Zhao, H.; Cornelius, L.A. Down-regulation of Rap1GAP via promoter hypermethylation promotes melanoma cell proliferation, survival, and migration. Cancer Res. 2009, 69, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Elola, M.T.; Wolfenstein-Todel, C.; Troncoso, M.F.; Vasta, G.R.; Rabinovich, G.A. Galectins: Matricellular glycan-binding proteins linking cell adhesion, migration, and survival. Cell. Mol. Life Sci. 2007, 64, 1679–1700. [Google Scholar] [CrossRef] [PubMed]
- Reinhold, M.I.; Lindberg, F.P.; Plas, D.; Reynolds, S.; Peters, M.G.; Brown, E.J. In vivo expression of alternatively spliced forms of integrin-associated protein (CD47). J. Cell Sci. 1995, 108, 3419–3425. [Google Scholar] [PubMed]
- Sadok, A.; Marshall, C. Rho GTPases: Masters of cell migration. Small GTPases 2014, 5, e29710. [Google Scholar] [CrossRef] [PubMed]
- Hsueh, Y. Calcium/calmodulin-dependent serine protein kinase and mental retardation. Ann. Neurol. 2009, 66, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Xing, F.; Okuda, H.; Watabe, M.; Kobayashi, A.; Pai, S.K.; Liu, W.; Pandey, P.R.; Fukuda, K.; Hirota, S.; Sugai, T.; et al. Hypoxia-induced Jagged2 promotes breast cancer metastasis and self-renewal of cancer stem-like cells. Oncogene 2011, 30, 4075–4086. [Google Scholar] [CrossRef] [PubMed]
- Van Rossum, A.G.; de Graaf, J.H.; Schuuring-Scholtes, E.; Kluin, P.M.; Fan, Y.X.; Zhan, X.; Moolenaar, W.H.; Schuuring, E. Alternative splicing of the actin binding domain of human cortactin affects cell migration. J. Biol. Chem. 2003, 278, 45672–45679. [Google Scholar] [CrossRef] [PubMed]
- Ch’ng, E.S.; Kumanogoh, A. Roles of Sema4D and Plexin-B1 in tumor progression. Mol. Cancer 2010, 9, 251. [Google Scholar] [CrossRef] [PubMed]
- David, C.J.; Manley, J.L. Alternative pre-mRNA splicing regulation in cancer: Pathways and programs unhinged. Genes Dev. 2010, 24, 2343–2364. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, X.; Wu, F.; Huang, R.; Xue, F.; Liang, G.; Tao, M.; Cai, P.; Huang, Y. Transcriptome profiling of the cancer, adjacent non-tumor and distant normal tissues from a colorectal cancer patient by deep sequencing. PLoS ONE 2012, 7, e41001. [Google Scholar] [CrossRef] [PubMed]
- Thorsen, K.; Sorensen, K.D.; Brems-Eskildsen, A.S.; Modin, C.; Gaustadnes, M.; Hein, A.M.; Kruhøffer, M.; Laurberg, S.; Borre, M.; Wang, K.; et al. Alternative splicing in colon, bladder, and prostate cancer identified by exon array analysis. Mol. Cell. Proteom. 2008, 7, 1214–1224. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, A.; Doci, C.L.; Gutkind, J.S. Semaphorins signaling in angiogenesis, lymphangiogenesis and cancer. Cell Res. 2012, 22, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Tamagnone, L. Emerging role of Semaphorins as major regulatory signals and potential therapeutic targets in cancer. Cancer Cell 2012, 22, 145–152. [Google Scholar] [CrossRef]
- Swiercz, J.; Worzfeld, T.; Offermans, S. ErbB-2 and met reciprocally regulate cellular signaling via plexin-B1. J. Biol. Chem. 2007, 283, 1893–1901. [Google Scholar] [CrossRef] [PubMed]
- Parker, M.W.; Hellman, L.M.; Xu, P.; Fried, M.G.; Vander Kooi, C.W. Furin processing of Semaphorin3F determines its anti-angiogenic activity by regulating direct binding and competition for neuropilin. Biochemistry 2010, 49, 4068–4075. [Google Scholar] [CrossRef] [PubMed]
- Doçi, C.L.; Mikelis, C.M.; Lionakis, M.S.; Molinolo, A.A.; Gutkind, J.S. Genetic identification of SEMA3F as an antilymphangiogenic metastasis suppressor gene in head and neck squamous carcinoma. Cancer Res. 2015, 75, 2937–2948. [Google Scholar] [CrossRef] [PubMed]
- Xiong, G.; Wang, C.; Evers, B.M.; Zhou, B.P.; Xu, R. RORα suppresses breast tumor invasion by inducing SEMA3F expression. Cancer Res. 2012, 72, 1728–1739. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef] [PubMed]
- Costa, V.; Esposito, R.; Ziviello, C.; Sepe, R.; Bim, L.V.; Cacciola, N.A.; Decaussin-Petrucci, M.; Pallante, P.; Fusco, A.; Ciccodicola, A. New somatic mutations and WNK1-B4GALNT3 gene fusion in papillary thyroid carcinoma. Oncotarget 2015, 6, 11242–11251. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-Seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aversa, R.; Sorrentino, A.; Esposito, R.; Ambrosio, M.R.; Amato, A.; Zambelli, A.; Ciccodicola, A.; D’Apice, L.; Costa, V. Alternative Splicing in Adhesion- and Motility-Related Genes in Breast Cancer. Int. J. Mol. Sci. 2016, 17, 121. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17010121
Aversa R, Sorrentino A, Esposito R, Ambrosio MR, Amato A, Zambelli A, Ciccodicola A, D’Apice L, Costa V. Alternative Splicing in Adhesion- and Motility-Related Genes in Breast Cancer. International Journal of Molecular Sciences. 2016; 17(1):121. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17010121
Chicago/Turabian StyleAversa, Rosanna, Anna Sorrentino, Roberta Esposito, Maria Rosaria Ambrosio, Angela Amato, Alberto Zambelli, Alfredo Ciccodicola, Luciana D’Apice, and Valerio Costa. 2016. "Alternative Splicing in Adhesion- and Motility-Related Genes in Breast Cancer" International Journal of Molecular Sciences 17, no. 1: 121. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17010121