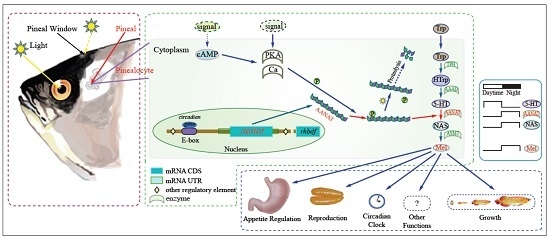
Molecular Evolution of Aralkylamine N-Acetyltransferase in Fish: A Genomic Survey
Abstract
:1. Introduction

2. Results
2.1. Variation of Aanat Copy Number in Different Species
| Class | Common Name | Species Name | aanat | Location | Accession Number | Source | |
|---|---|---|---|---|---|---|---|
| Nucleotide Sequence | Protein Sequence | ||||||
| Mammalis | Human | Homo sapiens | 1 | chromosome: 17 | NP_001160051.1 | EAW89400.1 | NCBI |
| House mouse | Mus musculus | 1 | chromosome: 11 | NM_009591.3 | EDL34585.1 | NCBI | |
| Chimpanzee | Pan troglodytes | 1 | chromosome: 17 | AY665273.2 | AAV74311.2 | NCBI | |
| Sheep | Ovis aries | 1 | chromosome: 11 | NM_001009461.1 | NP_001009461.1 | NCBI | |
| Minke whale | Balaenoptera acutorostrata scammoni | 1 | chromosome: Un | XM_007185968.1 | XP_007186030.1 | NCBI | |
| Platypus | Ornithorhynchus anatinus | 1 | Ultra430:237283–238471: −1 | ENSOANG00000011565 | ENSOANT00000018326 | Ensembl | |
| Panda | Ailuropoda melanoleuca | 1 | GL192703.1:1467291–1468587: 1 | ENSAMEG00000010321 | ENSAMET00000011309 | Ensembl | |
| Aves Reptilia | Rock pigeon | Columba livia | 1 | chromosome: Un | XM_005503150 | XP_005503207.1 | NCBI |
| Green sea turtle | Chelonia mydas | 1 | chromosome: Un | XM_007072534.1 | EMP23774 | NCBI | |
| Anole lizard | Anolis carolinensis | 1 | chromosome: 2 | XM_003217189.2 | XP_003217237.1 | NCBI | |
| Amphibia | Western clawed frog | Xenopus laevis | 1 | chromosome: Un | XM_002935933.2 | XP_002935979.1 | NCBI |
| Actinopterygii | Zebrafish | Danio rerio | 1 | chromosome: 6 | BC059448.1 | AAH59448.1 | NCBI |
| 2 | chromosome: 3 | NM_131411.2 | NP_571486.1 | NCBI | |||
| Mexican tetra | Astyanax mexicanus | 1 | KB882152.1:1429796–1431775: −1 | XM_007252167.1 | XP_007252229.1 | NCBI | |
| 2 | KB872051.1:338907-348651: −1 | ENSAMXG00000016495 | ENSAMXT00000016988 | Ensembl | |||
| Gilthead seabream | Sparus aurata | 1 | – | AY533402.1 | AAT02159.1 | NCBI | |
| 2 | – | AY533403.2 | AAT02160.2 | NCBI | |||
| European seabass | Dicentrarchus labrax | 1a | – | EU378922.1 | ACB13284.1 | NCBI | |
| 1b | – | EU378923.1 | ACB13285.1 | NCBI | |||
| Atlantic cod | Gadus morhua | 1 | GeneScaffold 1449:283672–285101: −1 | ENSGMOG00000010804 | ENSGMOT00000011867 | Ensembl | |
| 2 | GeneScaffold 1876:387650–388957: −1 | ENSGMOG00000004277 | ENSGMOT00000004660 | Ensembl | |||
| Medaka | Oryzias latipes | 1a | chromosome: 1 | NM_001104832.1 | NP_001098302.1 | NCBI | |
| 1b | chromosome: 8 | NM_001104860.1 | NP_001098330.1 | NCBI | |||
| 2 | chromosome: 8 | AB248276.1 | BAE78762.1 | NCBI | |||
| Fugu | Takifugu rubripes | 1a | scaffold 2391:5485–6279: −1 | ENSTRUG00000017502 | ENSTRUT00000045022 | Ensembl | |
| 1b | scaffold 65:466946-468376: −1 | ENSTRUG00000009927 | ENSTRUT00000025045 | Ensembl | |||
| 2 | scaffold15:451903–452747: −1 | ENSTRUG00000011652 | ENSTRUT00000029563 | Ensembl | |||
| Tilapia | Oreochromis niloticus | 1a | GL831193.1:3303726–3305445: 1 | ENSONIG00000018253 | ENSONIT00000023007 | Ensembl | |
| 1b | Un random:3751825–3761582: 1 | ENSTNIG00000004312 | ENSTNIT00000007096 | Ensembl | |||
| 2 | 3:12195390–12196528: −1 | ENSTNIG00000018581 | ENSONIT00000001810 | Ensembl | |||
| Tetraodon | Tetraodon nigroviridis | 1a | Un random:26969504–26970941: −1 | ENSTNIG00000017516 | ENSTNIT00000020893 | Ensembl | |
| 1b | Un random:3751825–3761582: 1 | ENSTNIG00000004312 | ENSTNIT00000007096 | Ensembl | |||
| 2 | 3:12195390–12196528: −1 | ENSTNIG00000018581 | ENSTNIT00000021990 | Ensembl | |||
| Amazon molly 1 | Poecilia formosa | 1a | KI519952.1:480412–483140: −1 | ENSPFOG00000012646 | ENSPFOT00000012663 | Ensembl | |
| 1b | KI519724.1:1037528–1042207: 1 | ENSPFOG00000003689 | ENSPFOT00000003582 | Ensembl | |||
| 2a | KI519725.1:742206-743398: −1 | ENSPFOG00000010360 | ENSPFOT00000010350 | Ensembl | |||
| 2b | KI519725.1:771388–772351:-1 | ENSPFOG00000010492 | ENSPFOT00000010477 | Ensembl | |||
| Platyfish | Xiphophorus maculatus | 1a | JH556678.1:1270978–1287717: 1 | ENSXMAG00000006015 | ENSXMAT00000006036 | Ensembl | |
| 1b | JH557102.1:45038–49228: 1 | ENSXMAG00000008291 | ENSXMAT00000008323 | Ensembl | |||
| 2 | JH556906.1:38852–39807: 1 | ENSXMAG00000000128 | ENSXMAT00000000129 | Ensembl | |||
| Stickleback 2 | Gasterosteus aculeatus | 1a | groupIX:16205994–16208003:-1 | ENSGACG00000019276 | ENSGACT00000025530 | Ensembl | |
| 1b | groupXI:3766738–3767762: −1 | ENSGACG00000007070 | ENSGACT00000009381 | Ensembl | |||
| 2 | groupXI:13702853–13703781: −1 | ENSGACG00000013957 | ENSGACT00000018464 | Ensembl | |||
| Chondrichthyes arcopterygii | Elephant shark | Callorhinchus milii | 1 | chromosome: Un | NM_001292772.1 | NP_001279701.1 | NCBI |
| Coelacanth | Latimeria chalumnae | 1 | chromosome: Un | XM_005993207.1 | XP_005993269.1 | NCBI | |
| Class | Common Name | Species Name | Total Number | aanat1a | aanat1b | aanat2 |
|---|---|---|---|---|---|---|
| Actinopterygii | Giant-fin mudskipper/PM 1 | Periophthalmus magnuspinnatus | 2 | – | 1 | 1 |
| Blue-spotted mudskipper/BP 1 | Boleophthalmus pectinirosris | 3 | 1 | 1 | 1 | |
| Malabar grouper | Epinephelus malabaricus | 3 | 1 | 1 | 1 | |
| Half-smooth tongue sole | Cynoglossus semilaevis | 3 | 1 | 1 | 1 | |
| Hedgehog seahorse | Hippocampus spinosissimus | 3 | 1 | 1 | 1 | |
| Atlantic salmon 2 | Salmo salar | 5 | 1 | 2 | 2 | |
| Rainbow trout 2 | Oncorhynchus mykiss | 5 | 1 | 2 | 2 | |
| Northern pike | Esox lucius | 3 | 1 | 1 | 1 | |
| Golden-line fishes (SIA) 2 | Sinocyclocheilus anshuiensis | 3 | 1 | – | 2 | |
| (SIG) 2 | Sinocyclocheilus graham | 3 | 1 | – | 2 | |
| (SIR) 2 | Sinocyclooheilus rhinocerous | 3 | 1 | – | 2 | |
| Asian arowana | Scleropages formosus | 3 | 1 | 1 | 1 |
2.2. The Phylogenetic Relationships among aanat Genes

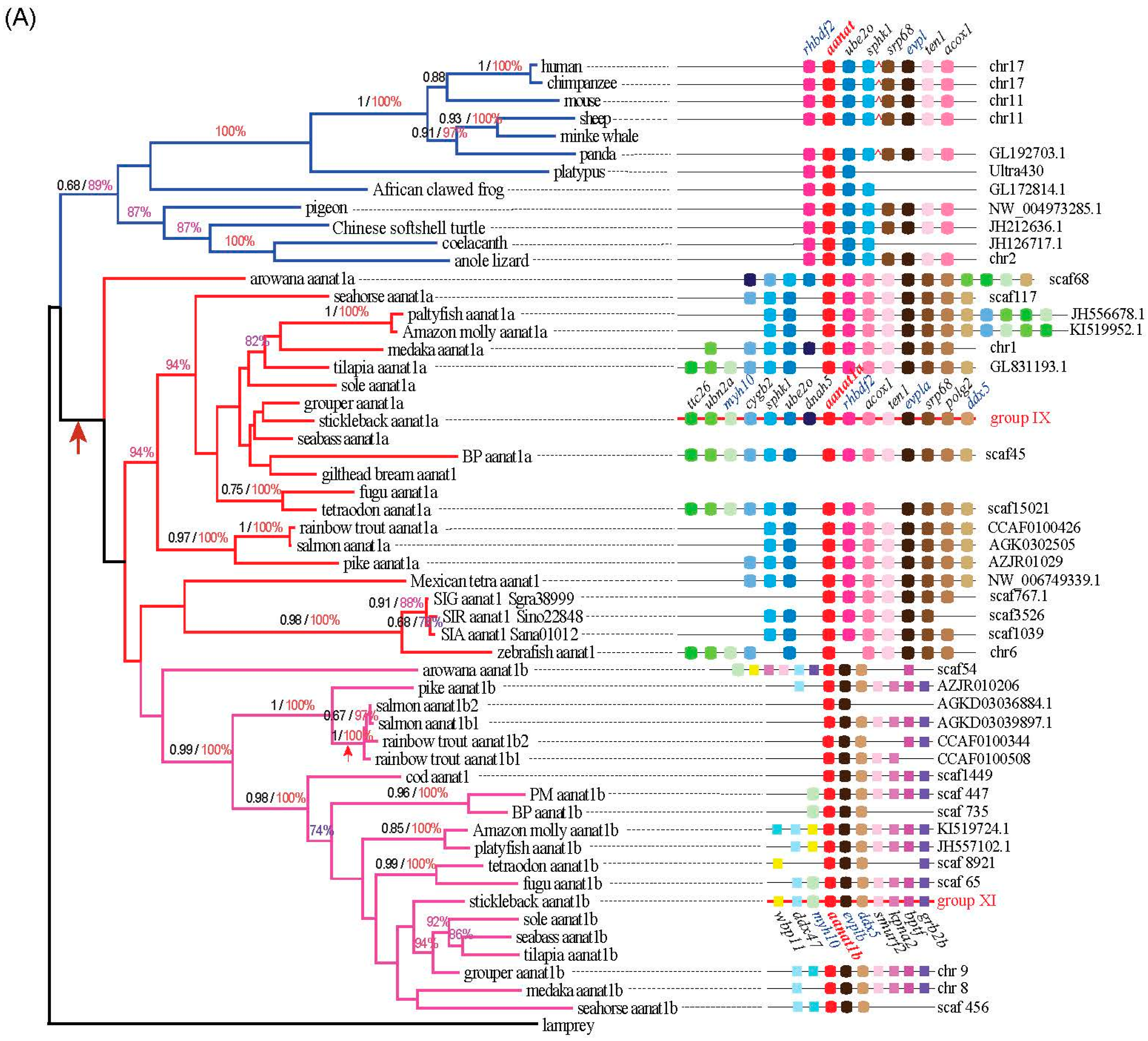
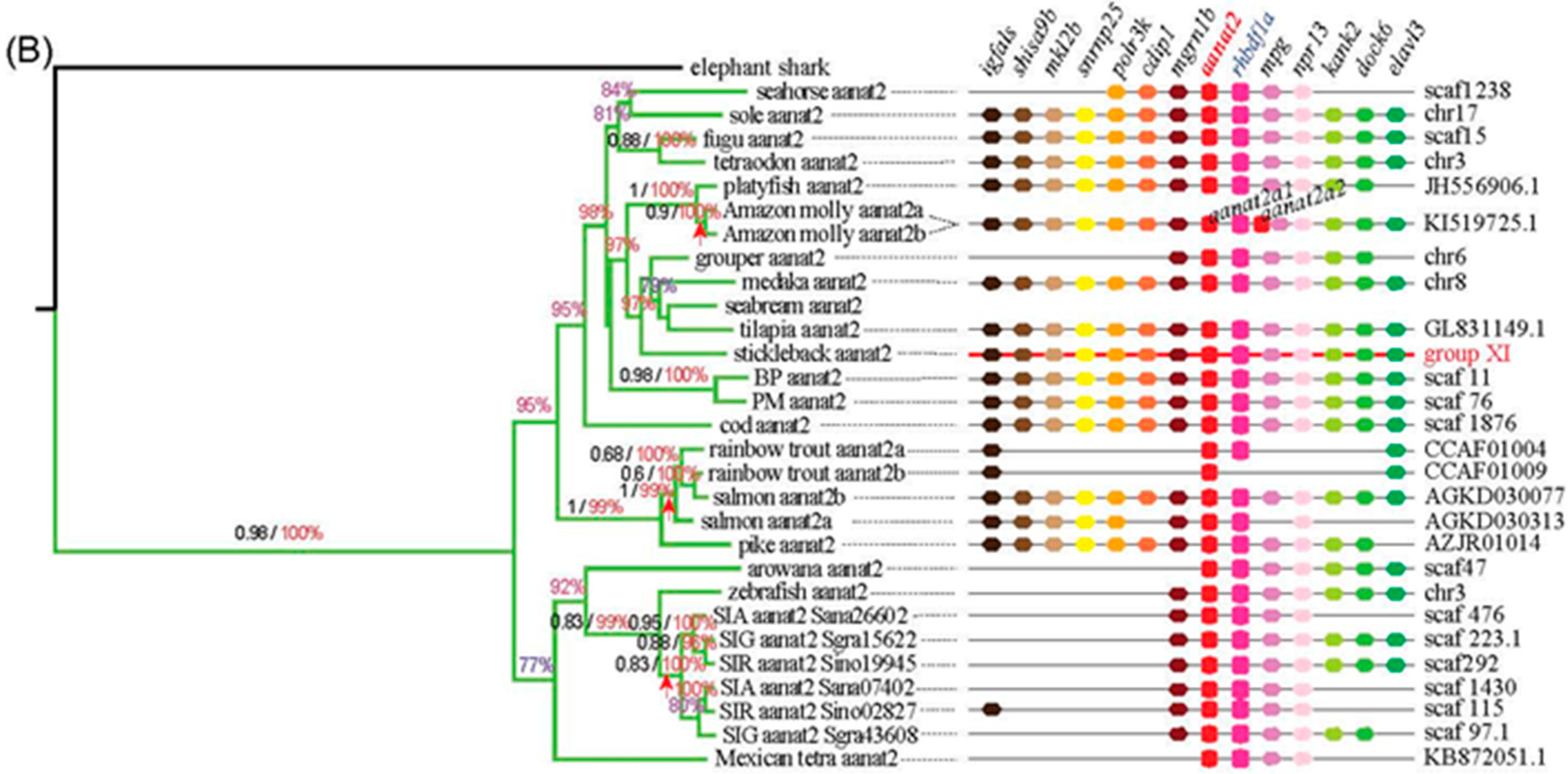
2.3. Synteny
2.4. The Regulatory Elements of aanat Genes
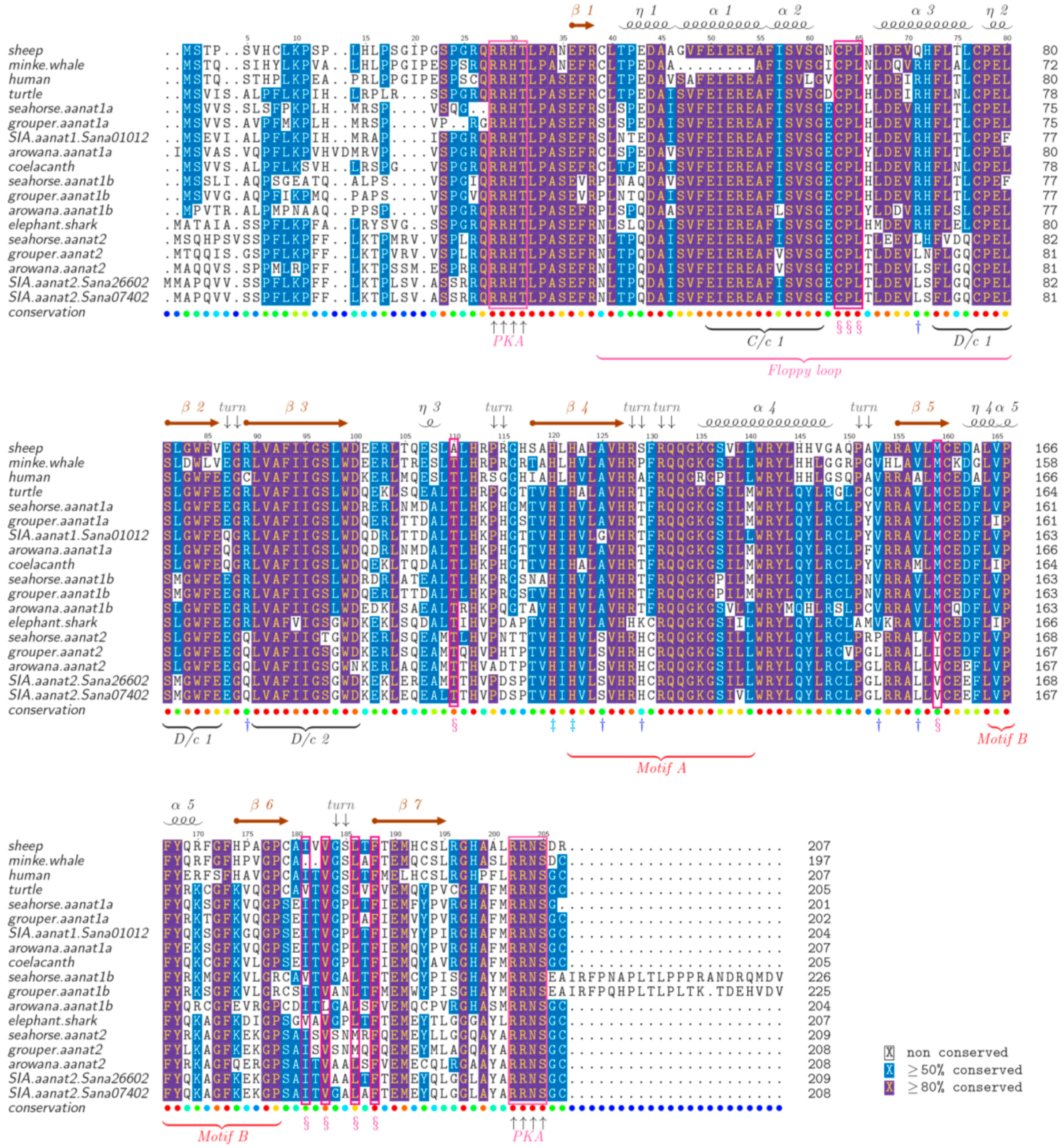

2.5. The Structure of AANAT Proteins
3. Discussion
3.1. The Potential Reasons of Copy Number Variation among Vertebrates
3.2. Adaptive Evolution of aanat
4. Experimental Section
4.1. Sequence Collection
4.2. Sequence Alignment and Phylogenetic Reconstruction
4.3. Identification of Conserved Synteny
4.4. Analysis of Regulatory Regions and the Differences between AANAT Proteins
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
References
- Iuvone, P.M.; Tosini, G.; Pozdeyev, N.; Haque, R.; Klein, D.C.; Chaurasia, S.S. Circadian clocks, clock networks, arylalkylamine n-acetyltransferase, and melatonin in the retina. Prog. Retin. Eye Res. 2005, 24, 433–456. [Google Scholar] [CrossRef] [PubMed]
- Li, D.Y.; Smith, D.G.; Hardeland, R.; Yang, M.Y.; Xu, H.L.; Zhang, L.; Yin, H.D.; Zhu, Q. Melatonin receptor genes in vertebrates. Int. J. Mol. Sci. 2013, 14, 11208–11223. [Google Scholar] [CrossRef] [PubMed]
- Hong, L.Y.; Hong, W.S.; Zhu, W.B.; Shi, Q.; You, X.X.; Chen, S.X. Cloning and expression of melatonin receptors in the mudskipper boleophthalmus pectinirostris: Their role in synchronizing its semilunar spawning rhythm. Gen. Comp. Endocrinol. 2014, 195, 138–150. [Google Scholar] [CrossRef] [PubMed]
- Pinillos, M.L.; De Pedro, N.; Alonso-Gomez, A.L.; Alonso-Bedate, M.; Delgado, M.J. Food intake inhibition by melatonin in goldfish (Carassius auratus). Physiol. Behav. 2001, 72, 629–634. [Google Scholar] [CrossRef]
- Lopez Patino, M.A.; Guijarro, A.I.; Alonso-Gomez, A.L.; Delgado, M.J. Characterization of two different melatonin binding sites in peripheral tissues of the teleost tinca tinca. Gen. Comp. Endocrinol. 2012, 175, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Velarde, E.; Alonso-Gomez, A.L.; Azpeleta, C.; Isorna, E.; Delgado, M.J. Melatonin attenuates the acetylcholine-induced contraction in isolated intestine of a teleost fish. J. Comp. Physiol. B 2009, 179, 951–959. [Google Scholar] [CrossRef] [PubMed]
- Lima-Cabello, E.; Diaz-Casado, M.E.; Guerrero, J.A.; Otalora, B.B.; Escames, G.; Lopez, L.C.; Reiter, R.J.; Acuna-Castroviejo, D. A review of the melatonin functions in zebrafish physiology. J. Pineal Res. 2014, 57, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hassell, K.J.; Reiter, R.J.; Robertson, N.J. Melatonin and its role in neurodevelopment during the perinatal period: A review. Fetal Matern. Med. Rev. 2013, 24, 76–107. [Google Scholar] [CrossRef]
- Lombardo, F.; Gioacchini, G.; Fabbrocini, A.; Candelma, M.; D’Adamo, R.; Giorgini, E.; Carnevali, O. Melatonin-mediated effects on killifish reproductive axis. Comp. Biochem Physiol. Part A 2014, 172, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Appelbaum, L.; Wang, G.X.; Maro, G.S.; Mori, R.; Tovin, A.; Marin, W.; Yokogawa, T.; Kawakami, K.; Smith, S.J.; Gothilf, Y.; et al. Sleep-wake regulation and hypocretin-melatonin interaction in zebrafish. Proc. Natl. Acad. Sci. USA 2009, 106, 21942–21947. [Google Scholar] [CrossRef] [PubMed]
- Klein, D.C.; Coon, S.L.; Roseboom, P.H.; Weller, J.L.; Bernard, M.; Gastel, J.A.; Zatz, M.; Iuvone, P.M.; Rodriguez, I.R.; Begay, V.; et al. The melatonin rhythm-generating enzyme: Molecular regulation of serotonin n-acetyltransferase in the pineal gland. Recent Prog. Horm. Res. 1997, 52, 307–357. [Google Scholar] [PubMed]
- Klein, D.C. Arylalkylamine n-acetyltransferase: “The timezyme”. J. Biol. Chem. 2007, 282, 4233–4237. [Google Scholar] [CrossRef] [PubMed]
- Cazamea-Catalan, D.; Besseau, L.; Falcon, J.; Magnanou, E. The timing of timezyme diversification in vertebrates. PLoS ONE 2014, 9, e112380. [Google Scholar] [CrossRef] [PubMed]
- Falcon, J.; Coon, S.L.; Besseau, L.; Cazamea-Catalan, D.; Fuentes, M.; Magnanou, E.; Paulin, C.H.; Boeuf, G.; Sauzet, S.; Jorgensen, E.H.; et al. Drastic neofunctionalization associated with evolution of the timezyme aanat 500 mya. Proc. Natl. Acad. Sci. USA 2014, 111, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Pavlicek, J.; Sauzet, S.; Besseau, L.; Coon, S.L.; Weller, J.L.; Boeuf, G.; Gaildrat, P.; Omelchenko, M.V.; Koonin, E.V.; Falcon, J.; et al. Evolution of aanat: Expansion of the gene family in the cephalochordate amphioxus. BMC Evol. Biol. 2010, 10, 154. [Google Scholar] [CrossRef] [PubMed]
- Appelbaum, L.; Vallone, D.; Anzulovich, A.; Ziv, L.; Tom, M.; Foulkes, N.S.; Gothilf, Y. Zebrafish arylalkylamine-n-acetyltransferase genes-targets for regulation of the circadian clock. J. Mol. Endocrinol. 2006, 36, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Appelbaum, L.; Toyama, R.; Dawid, I.B.; Klein, D.C.; Baler, R.; Gothilf, Y. Zebrafish serotonin-n-acetyltransferase-2 gene regulation: Pineal-restrictive downstream module contains a functional e-box and three photoreceptor conserved elements. Mol. Endocrinol. 2004, 18, 1210–1221. [Google Scholar] [CrossRef] [PubMed]
- Tosini, G.; Chaurasia, S.S.; Michael Iuvone, P. Regulation of arylalkylamine n-acetyltransferase (aanat) in the retina. Chronobiol. Int. 2006, 23, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, S.; Gastel, J.A.; Weller, J.L.; Schwartz, C.; Jaffe, H.; Namboodiri, M.A.; Coon, S.L.; Hickman, A.B.; Rollag, M.; Obsil, T.; et al. Role of a pineal camp-operated arylalkylamine n-acetyltransferase/14-3-3-binding switch in melatonin synthesis. Proc. Natl. Acad. Sci. USA 2001, 98, 8083–8088. [Google Scholar] [CrossRef] [PubMed]
- Asaoka, Y.; Mano, H.; Kojima, D.; Fukada, Y. Pineal expression-promoting element (pipe), a cis-acting element, directs pineal-specific gene expression in zebrafish. Proc. Natl. Acad. Sci. USA 2002, 99, 15456–15461. [Google Scholar] [CrossRef] [PubMed]
- Falcon, J.; Galarneau, K.M.; Weller, J.L.; Ron, B.; Chen, G.; Coon, S.L.; Klein, D.C. Regulation of arylalkylamine n-acetyltransferase-2 (aanat2, ec 2.3.1.87) in the fish pineal organ: Evidence for a role of proteasomal proteolysis. Endocrinology 2001, 142, 1804–1813. [Google Scholar] [CrossRef] [PubMed]
- Cazamea-Catalan, D.; Magnanou, E.; Helland, R.; Besseau, L.; Boeuf, G.; Falcon, J.; Jorgensen, E.H. Unique arylalkylamine n-acetyltransferase-2 polymorphism in salmonids and profound variations in thermal stability and catalytic efficiency conferred by two residues. J. Exp. Biol. 2013, 216, 1938–1948. [Google Scholar] [CrossRef] [PubMed]
- Falcon, J.; Migaud, H.; Munoz-Cueto, J.A.; Carrillo, M. Current knowledge on the melatonin system in teleost fish. Gen. Comp. Endocrinol. 2010, 165, 469–482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gandhi, A.V.; Mosser, E.A.; Oikonomou, G.; Prober, D.A. Melatonin is required for the circadian regulation of sleep. Neuron 2015, 85, 1193–1199. [Google Scholar] [CrossRef] [PubMed]
- Paulin, C.H.; Cazamea-Catalan, D.; Zilberman-Peled, B.; Herrera-Perez, P.; Sauzet, S.; Magnanou, E.; Fuentes, M.; Gothilf, Y.; Munoz-Cueto, J.A.; Falcon, J.; et al. Subfunctionalization of arylalkylamine n-acetyltransferases in the sea bass dicentrarchus labrax: Two-ones for one two. J. Pineal Res. 2015, 59, 354–364. [Google Scholar] [CrossRef] [PubMed]
- Zilberman-Peled, B.; Ron, B.; Gross, A.; Finberg, J.P.; Gothilf, Y. A possible new role for fish retinal serotonin-n-acetyltransferase-1 (aanat1): Dopamine metabolism. Brain Res. 2006, 1073–1074, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Nisembaum, L.G.; Tinoco, A.B.; Moure, A.L.; Alonso Gomez, A.L.; Delgado, M.J.; Valenciano, A.I. The arylalkylamine-n-acetyltransferase (aanat) acetylates dopamine in the digestive tract of goldfish: A role in intestinal motility. Neurochem. Int. 2013, 62, 873–880. [Google Scholar] [CrossRef] [PubMed]
- Velarde, E.; Cerda-Reverter, J.M.; Alonso-Gomez, A.L.; Sanchez, E.; Isorna, E.; Delgado, M.J. Melatonin-synthesizing enzymes in pineal, retina, liver, and gut of the goldfish (carassius): mRNA expression pattern and regulation of daily rhythms by lighting conditions. Chronobiol. Int. 2010, 27, 1178–1201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coon, S.L.; Klein, D.C. Evolution of arylalkylamine n-acetyltransferase: Emergence and divergence. Mol. Cell. Endocrinol. 2006, 252, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Scheibner, K.A.; De Angelis, J.; Burley, S.K.; Cole, P.A. Investigation of the roles of catalytic residues in serotonin n-acetyltransferase. J. Biol. Chem. 2002, 277, 18118–18126. [Google Scholar] [CrossRef] [PubMed]
- Hickman, A.B.; Klein, D.C.; Dyda, F. Melatonin biosynthesis: The structure of serotonin n-acetyltransferase at 2.5 a resolution suggests a catalytic mechanism. Mol. Cell 1999, 3, 23–32. [Google Scholar] [CrossRef]
- Guyomard, R.; Boussaha, M.; Krieg, F.; Hervet, C.; Quillet, E. A synthetic rainbow trout linkage map provides new insights into the salmonid whole genome duplication and the conservation of synteny among teleosts. BMC Genet. 2012, 13, 15. [Google Scholar] [CrossRef] [PubMed]
- Glasauer, S.M.; Neuhauss, S.C. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genom. 2014, 289, 1045–1060. [Google Scholar] [CrossRef] [PubMed]
- Zilberman-Peled, B.; Bransburg-Zabary, S.; Klein, D.C.; Gothilf, Y. Molecular evolution of multiple arylalkylamine n-acetyltransferase (aanat) in fish. Mar. Drugs 2011, 9, 906–921. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, O.; Aury, J.M.; Brunet, F.; Petit, J.L.; Stange-Thomann, N.; Mauceli, E.; Bouneau, L.; Fischer, C.; Ozouf-Costaz, C.; Bernot, A.; et al. Genome duplication in the teleost fish tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 2004, 431, 946–957. [Google Scholar] [CrossRef] [PubMed]
- Berthelot, C.; Brunet, F.; Chalopin, D.; Juanchich, A.; Bernard, M.; Noel, B.; Bento, P.; Da Silva, C.; Labadie, K.; Alberti, A.; et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat. Commun. 2014, 5, 3657. [Google Scholar] [CrossRef] [PubMed]
- Shotgun Assembly Sequences: Genome (WGS) and Transcriptome (TSA). Available online: http://0-www-Ncbi-Nlm-Nih-Gov.brum.beds.ac.uk/traces/wgs/?val=CBXY01 (accessed on 5 March 2015).
- Isorna, E.; Aliaga-Guerrero, M.; M’Rabet, A.E.; Servili, A.; Falcon, J.; Munoz-Cueto, J.A. Identification of two arylalkylamine n-acetyltranferase 1 genes with different developmental expression profiles in the flatfish solea senegalensis. J. Pineal Res. 2011, 51, 434–444. [Google Scholar] [CrossRef]
- You, X.; Bian, C.; Zan, Q.; Xu, X.; Liu, X.; Chen, J.; Wang, J.; Qiu, Y.; Li, W.; Zhang, X.; et al. Mudskipper genomes provide insights into the terrestrial adaptation of amphibious fishes. Nat. Commun. 2014, 5, 5594. [Google Scholar] [CrossRef] [PubMed]
- Feldkaemper, M.; Schaeffel, F. An updated view on the role of dopamine in myopia. Exp. Eye Res. 2013, 114, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Mount, D.W. Using the basic local alignment search tool (blast). CSH Protoc. 2004, 2007. [Google Scholar] [CrossRef] [PubMed]
- Birney, E.; Durbin, R. Using genewise in the drosophila annotation experiment. Genom. Res. 2000, 10, 547–548. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. Mafft: A novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Available online: http://phylogeny.Lirmm.Fr/ (accessed on 19 April 2008).
- Mrmtgui. V 1.0; Paulo Nuin: Boston, MA, USA, 2007.
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Huggins, P.M.; Li, W.; Haws, D.; Friedrich, T.; Liu, J.; Yoshida, R. Bayes estimators for phylogenetic reconstruction. Syst. Biol. 2011, 60, 528–540. [Google Scholar] [CrossRef] [PubMed]
- Criscuolo, A. Morephyml: Improving the phylogenetic tree space exploration with phyml 3. Mol. Phylogenet. Evol. 2011, 61, 944–948. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. Mega6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. Prottest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [PubMed]
- Beitz, E. Texshade: Shading and labeling of multiple sequence alignments using latex2 epsilon. Bioinformatics 2000, 16, 135–139. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; You, X.; Bian, C.; Yu, H.; Coon, S.L.; Shi, Q. Molecular Evolution of Aralkylamine N-Acetyltransferase in Fish: A Genomic Survey. Int. J. Mol. Sci. 2016, 17, 51. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17010051
Li J, You X, Bian C, Yu H, Coon SL, Shi Q. Molecular Evolution of Aralkylamine N-Acetyltransferase in Fish: A Genomic Survey. International Journal of Molecular Sciences. 2016; 17(1):51. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17010051
Chicago/Turabian StyleLi, Jia, Xinxin You, Chao Bian, Hui Yu, Steven L. Coon, and Qiong Shi. 2016. "Molecular Evolution of Aralkylamine N-Acetyltransferase in Fish: A Genomic Survey" International Journal of Molecular Sciences 17, no. 1: 51. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17010051