Assessing Heterogeneity of Osteolytic Lesions in Multiple Myeloma by 1H HR-MAS NMR Metabolomics
Abstract
:1. Introduction
2. Results
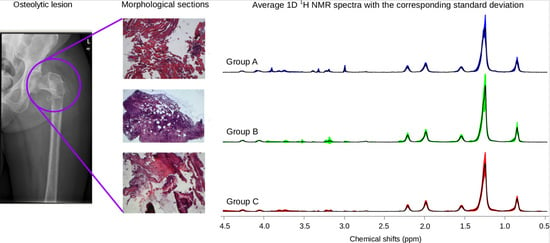
2.1. 1H HR-MAS Spectra Highlight Differences among Multiple Biopsy Specimens
2.2. Sample Recovery from HR-MAS Rotors Allows Histology and Histomorphometry Analysis
2.3. Unsupervised Multivariate Statistical Analysis of 1H HR-MAS NMR Data
2.4. Integrated Analysis of Histomorphometric Features and NMR Metabolic Profiles
3. Discussion
4. Materials and Methods
4.1. Case History and Sampling
4.2. Sample Preparation for NMR
4.3. 1H HR-MAS NMR
4.4. Binning and Profiling
4.5. Statistical Analyses of NMR Data
4.5.1. Univariate Analysis
4.5.2. Multivariate Analysis
4.6. Histology and Histomorphometry
4.7. Correlation Analyses
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Palumbo, A.; Anderson, K. Multiple myeloma. N. Engl. J. Med. 2011, 364, 1046–1060. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, S.V.; Merlini, G.; San Miguel, J.F. Haematological cancer: Redefining myeloma. Nat. Rev. Clin. Oncol. 2012, 9, 494–496. [Google Scholar] [CrossRef] [PubMed]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; Tarpey, P.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Ramautar, R.; Berger, R.; van der Greef, J.; Hankemeier, T. Human metabolomics: strategies to understand biology. Curr. Opin. Chem. Biol. 2013, 17, 841–846. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Holmes, E.; Kinross, J.M.; Darzi, A.W.; Takats, Z.; Lindon, J.C. Metabolic phenotyping in clinical and surgical environments. Nature 2012, 491, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Bathen, T.F.; Geurts, B.; Sitterm, B.; Fjosne, H.E.; Lundgren, S.; Buydens, L.M.; Gribbestad, I.S.; Postma, G.; Giskeødegård, G.F. Feasibility of MR metabolomics for immediate analysis of resection margins during breast cancer surgery. PLoS ONE 2013, 8, e61578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beckonert, O.; Coen, M.; Keun, H.C.; Wang, Y.; Ebbels, T.M.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nat. Protoc. 2010, 5, 1019–1032. [Google Scholar] [CrossRef] [PubMed]
- Moestue, S.; Sitter, B.; Bathen, T.F.; Tessem, M.B.; Gribbestad, I.S. HR MAS MR spectroscopy in metabolic characterization of cancer. Curr. Top. Med. Chem. 2011, 11, 2–26. [Google Scholar] [CrossRef] [PubMed]
- Piotto, M.; Moussallieh, F.M.; Euville, A.; Bellocq, J.P.; Elbayed, K.; Namer, I.J. Towards real-time metabolic profiling of a biopsy specimen N during a surgical operation by 1H high resolution magic angle spinning nuclear magnetic resonance: A case report. J. Med. Case Rep. 2012, 6, 22. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.F.; Kurhanewicz, J.; Tabatabai, Z.L.; Simko, J.P.; Keshari, K.R.; Gbegnon, A.; Santos, R.D.; Federman, S.; Shinohara, K.; Carroll, P.R.; et al. Metabolic, pathologic, and genetic analysis of prostate tissues: quantitative evaluation of histopathologic and mRNA integrity after HR-MAS spectroscopy. NMR Biomed. 2010, 23, 391–398. [Google Scholar] [PubMed]
- Tzika, A.A.; Astrakas, L.; Cao, H.; Mintzopoulos, D.; Andronesi, O.C.; Mindrinos, M.; Zhang, J.; Rahme, L.G.; Blekas, K.D.; Likas, A.C. Combination of high-resolution magic angle spinning proton magnetic resonance spectroscopy and microscale genomics to type brain tumor biopsies. Int. J. Mol. Med. 2007, 20, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Righi, V.; Roda, J.M.; Paz, J.; Mucci, A.; Tugnoli, V.; Rodriguez-Tarduchy, G.; Barrios, L.; Schenetti, L.; Cerdán, S.; García-Martín, M.L. 1H HR-MAS and genomic analysis of human tumor biopsies discriminate between high and low grade astrocytomas. NMR Biomed. 2009, 22, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.L.; Anthony, D.C.; Comite, A.R.; Black, P.M.; Tzika, A.A.; Gonzalez, R.G. Quantification of microheterogeneity in glioblastoma multiforme with ex vivo high-resolution magic-angle spinning (HRMAS) proton magnetic resonance spectroscopy. Neuro Oncol. 2000, 2, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Sjobakk, T.E.; Vettukattil, R.; Gulati, M.; Gulati, S.; Lundgren, S.; Gribbestad, I.S.; Torp, S.H.; Bathen, T.F. Metabolic profiles of brain metastases. Int. J. Mol. Sci. 2013, 14, 2104–2118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Righi, V.; Mucci, A.; Schenetti, L.; Bacci, A.; Agati, R.; Leonardi, M.; Schiavina, R.; Martorana, G. Identification of mobile lipids in human cancer tissues by ex vivo diffusion edited HR-MAS MRS. Oncol. Rep. 2009, 22, 1493–1496. [Google Scholar] [PubMed]
- Bathen, T.F.; Sitter, B.; Sjobakk, T.E.; Tessem, M.B.; Gribbestad, I.S. Magnetic resonance metabolomics of intact tissue: a biotechnological tool in cancer diagnostics and treatment evaluation. Cancer Res. 2010, 70, 6692–6696. [Google Scholar] [CrossRef] [PubMed]
- Duarte, I.F.; Stanley, E.G.; Holmes, E.; Lindon, J.C.; Gil, A.M.; Tang, H.; Ferdinand, R.; McKee, C.G.; Nicholson, J.K.; Vilca-Melendez, H.; et al. Metabolic assessment of human liver transplants from biopsy samples at the donor and recipient stages using high-resolution magic angle spinning 1H NMR spectroscopy. Anal. Chem. 2005, 77, 5570–5578. [Google Scholar] [CrossRef] [PubMed]
- Mirnezami, R.; Jimenez, B.; Li, J.V.; Kinross, J.M.; Veselkov, K.; Goldin, R.D.; Holmes, E.; Nicholson, J.K.; Darzi, A. Rapid Diagnosis and Staging of Colorectal Cancer via High-Resolution Magic Angle Spinning Nuclear Magnetic Resonance (HR-MAS NMR) Spectroscopy of Intact Tissue Biopsies. Ann. Surg. 2014, 259, 1138–1149. [Google Scholar] [CrossRef] [PubMed]
- Thysell, E.; Surowiec, I.; Hornberg, E.; Crnalic, S.; Widmark, A.; Johansson, A.I.; Stattin, P.; Bergh, A.; Moritz, T.; Antti, H.; et al. Metabolomic characterization of human prostate cancer bone metastases reveals increased levels of cholesterol. PLoS ONE 2010, 5, e14175. [Google Scholar] [CrossRef] [PubMed]
- Borel, M.; Pastoureau, P.; Papon, J.; Madelmont, J.C.; Moins, N.; Maublant, J.; Miot-Noirault, E. Longitudinal profiling of articular cartilage degradation in osteoarthritis by high-resolution magic angle spinning 1H NMR spectroscopy: Experimental study in the meniscectomized guinea pig model. J. Proteome Res. 2009, 8, 2594–2600. [Google Scholar] [CrossRef] [PubMed]
- Keshari, K.R.; Lotz, J.C.; Kurhanewicz, J.; Majumdar, S. Correlation of HR-MAS spectroscopy derived metabolite concentrations with collagen and proteoglycan levels and Thompson grade in the degenerative disc. Spine 2005, 30, 2683–2688. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Hu, J.Z.; Rommereim, D.N.; Murphy, M.K.; Phipps, R.P.; Huso, D.L.; Dicello, J.F. Application of high-resolution 1H MAS NMR spectroscopy to the analysis of intact bones from mice exposed to gamma radiation. Radiat. Res. 2009, 172, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Kaflak-Hachulska, A.; Samoson, A.; Kolodziejski, W. 1H MAS and 1H→31P CP/MAS NMR study of human bone mineral. Calcif. Tissue Int. 2003, 73, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Song, Y.; Cho, N.; Chang, J.M.; Koo, H.R.; Yi, A.; Kim, H.; Park, S.; Moon, W.K. An HR-MAS MR metabolomics study on breast tissues obtained with core needle biopsy. PLoS ONE 2011, 6, e25563. [Google Scholar] [CrossRef] [PubMed]
- Lindon, J.C.; Beckonert, O.P.; Holmes, E.; Nicholson, J.K. High Resolution Magic Angle Spinning NMR Spetroscopy: Application to biomedical studies. Prog. Nucl. Magn. Res. 2009, 55, 79–100. [Google Scholar] [CrossRef]
- Shet, K.; Siddiqui, S.M.; Yoshihara, H.; Kurhanewicz, J.; Ries, M.; Li, X. High-resolution magic angle spinning NMR spectroscopy of human osteoarthritic cartilage. NMR Biomed. 2012, 25, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Righi, V.; Durante, C.; Cocchi, M.; Calabrese, C.; Di Febo, G.; Lecce, F.; Pisi, A.; Tugnoli, V.; Mucci, A.; Schenetti, L. Discrimination of healthy and neoplastic human colon tissues by ex vivo HR-MAS NMR spectroscopy and chemometric analyses. J. Proteome Res. 2009, 8, 1859–1869. [Google Scholar] [CrossRef] [PubMed]
- Fauvelle, F.; Carpentier, P.; Dorandeu, F.; Foquin, A.; Testylier, G. Prediction of neuroprotective treatment efficiency using a HRMAS NMR-based statistical model of refractory status epilepticus on mouse: A metabolomic approach supported by histology. J. Proteome Res. 2012, 11, 3782–3795. [Google Scholar] [CrossRef] [PubMed]
- Bouwman, J.; Vogels, J.T.; Wopereis, S.; Rubingh, C.M.; Bijlsma, S.; Ommen, B. Visualization and identification of health space, based on personalized molecular phenotype and treatment response to relevant underlying biological processes. BMC Med. Genom. 2012, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Righi, V.; Mucci, A.; Schenetti, L.; Tosi, M.R.; Grigioni, W.F.; Corti, B.; Bertaccini, A.; Franceschelli, A.; Sanguedolce, F.; Schiavina, R.; et al. Ex vivo HR-MAS magnetic resonance spectroscopy of normal and malignant human renal tissues. Anticancer Res. 2007, 27, 3195–3204. [Google Scholar] [PubMed]
- Opstad, K.S.; Bell, B.A.; Griffiths, J.R.; Howe, F.A. An investigation of human brain tumour lipids by high-resolution magic angle spinning 1H MRS and histological analysis. NMR Biomed. 2008, 21, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Benahmed, M.A.; Santelmo, N.; Elbayed, K.; Frossard, N.; Noll, E.; Canuet, M.; Pottecher, J.; Diemunsch, P.; Piotto, M.; Massard, G.; et al. The assessment of the quality of the graft in an animal model for lung transplantation using the metabolomics 1H high-resolution magic angle spinning NMR spectroscopy. Magn. Reson. Med. 2012, 68, 1026–1038. [Google Scholar] [CrossRef] [PubMed]
- Monleón, D.; Morales, J.M.; Gonzalez-Darder, J.; Talamantes, F.; Cortés, O.; Gil-Benso, R.; López-Ginés, C.; Cerdá-Nicolás, M.; Celda, B. Benign and atypical meningioma metabolic signatures by high-resolution magic-angle spinning molecular profiling. J. Proteome Res. 2008, 7, 2882–2888. [Google Scholar] [CrossRef] [PubMed]
- Chae, E.Y.; Shin, H.J.; Kim, S.; Baek, H.M.; Yoon, D.; Kim, S.; Shim, Y.E.; Kim, H.H.; Cha, J.H.; Choi, W.J.; et al. The Role of High-Resolution Magic Angle Spinning 1H Nuclear Magnetic Resonance Spectroscopy for Predicting the Invasive Component in Patients with Ductal Carcinoma In Situ Diagnosed on Preoperative Biopsy. PLoS ONE 2016, e0161038. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Mandal, R.; Sinelnikov, I.V.; Broadhurst, D.; Wishart, D.S. MetaboAnalyst 2.0—A comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012, 40, W127–W133. [Google Scholar] [CrossRef] [PubMed]





© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tavel, L.; Fontana, F.; Garcia Manteiga, J.M.; Mari, S.; Mariani, E.; Caneva, E.; Sitia, R.; Camnasio, F.; Marcatti, M.; Cenci, S.; et al. Assessing Heterogeneity of Osteolytic Lesions in Multiple Myeloma by 1H HR-MAS NMR Metabolomics. Int. J. Mol. Sci. 2016, 17, 1814. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17111814
Tavel L, Fontana F, Garcia Manteiga JM, Mari S, Mariani E, Caneva E, Sitia R, Camnasio F, Marcatti M, Cenci S, et al. Assessing Heterogeneity of Osteolytic Lesions in Multiple Myeloma by 1H HR-MAS NMR Metabolomics. International Journal of Molecular Sciences. 2016; 17(11):1814. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17111814
Chicago/Turabian StyleTavel, Laurette, Francesca Fontana, Josè Manuel Garcia Manteiga, Silvia Mari, Elisabetta Mariani, Enrico Caneva, Roberto Sitia, Francesco Camnasio, Magda Marcatti, Simone Cenci, and et al. 2016. "Assessing Heterogeneity of Osteolytic Lesions in Multiple Myeloma by 1H HR-MAS NMR Metabolomics" International Journal of Molecular Sciences 17, no. 11: 1814. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17111814