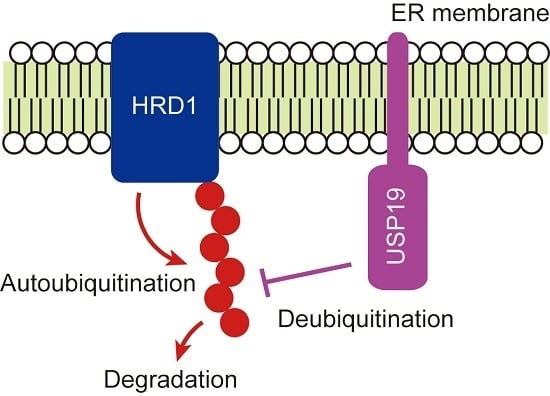
USP19-Mediated Deubiquitination Facilitates the Stabilization of HRD1 Ubiquitin Ligase
Abstract
:1. Introduction
2. Results
2.1. USP19 Interacts with HRD1
2.2. USP19 Deubiquitinates HRD1
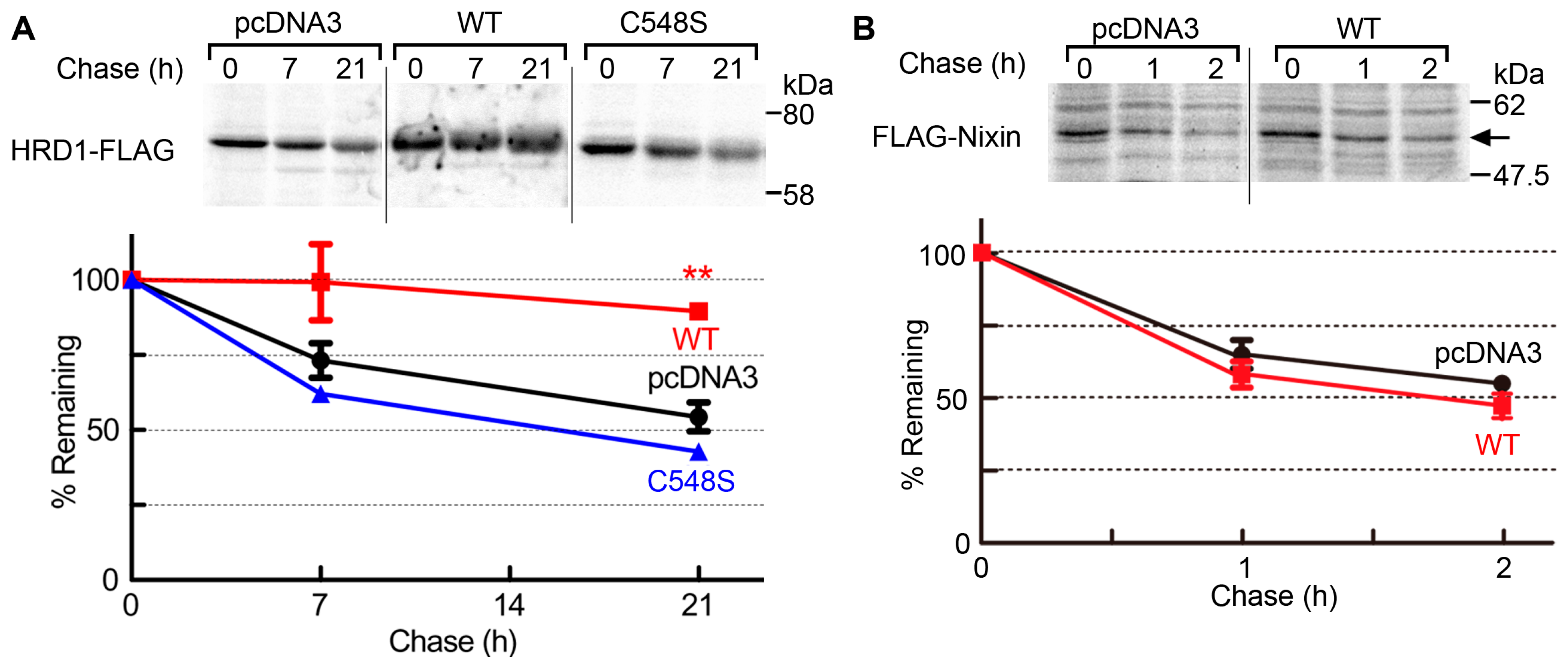
2.3. USP19 Stabilizes HRD1
3. Discussion
4. Materials and Methods
4.1. Plasmid Construction
4.2. Antibodies
4.3. Cell Culture and Plasmid Transfection
4.4. RNA Interference
4.5. Western Blotting and Immunoprecipitation
4.6. Pulse-Chase Experiments
4.7. Reverse Transcriptase (RT)-PCR
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fraile, J.M.; Quesada, V.; Rodriguez, D.; Freije, J.M.; Lopez-Otin, C. Deubiquitinases in cancer: New functions and therapeutic options. Oncogene 2012, 31, 2373–2388. [Google Scholar] [CrossRef] [PubMed]
- Hassink, G.C.; Zhao, B.; Sompallae, R.; Altun, M.; Gastaldello, S.; Zinin, N.V.; Masucci, M.G.; Lindsten, K. The ER-resident ubiquitin-specific protease 19 participates in the UPR and rescues ERAD substrates. EMBO Rep. 2009, 10, 755–761. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, N.; Hirose, S. Regulation of mitochondrial morphology by USP30, a deubiquitinating enzyme present in the mitochondrial outer membrane. Mol. Biol. Cell 2008, 19, 1903–1911. [Google Scholar] [CrossRef] [PubMed]
- Bedard, N.; Jammoul, S.; Moore, T.; Wykes, L.; Hallauer, P.L.; Hastings, K.E.; Stretch, C.; Baracos, V.; Chevalier, S.; Plourde, M.; et al. Inactivation of the ubiquitin-specific protease 19 deubiquitinating enzyme protects against muscle wasting. FASEB J. 2015, 29, 3889–3898. [Google Scholar] [CrossRef] [PubMed]
- Combaret, L.; Adegoke, O.A.; Bedard, N.; Baracos, V.; Attaix, D.; Wing, S.S. USP19 is a ubiquitin-specific protease regulated in rat skeletal muscle during catabolic states. Am. J. Physiol. Endocrinol. Metab. 2005, 288, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Xu, W.G.; Luo, Y.; Han, F.F.; Yao, X.H.; Yang, T.Y.; Zhang, Y.; Pi, W.F.; Guo, X.J. Cigarette smoke-induced skeletal muscle atrophy is associated with up-regulation of USP-19 via p38 and ERK MAPKs. J. Cell. Biochem. 2011, 112, 2307–2316. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Kariya, Y.; Kitakaze, T.; Yamaji, R.; Harada, N.; Sakamoto, T.; Hosotani, K.; Nakano, Y.; Inui, H. The preventive effect of β-carotene on denervation-induced soleus muscle atrophy in mice. Br. J. Nutr. 2013, 109, 1349–1358. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, P.; Pang, Z.; Miao, M.; Yu, L.; Wing, S.S. USP19-deubiquitinating enzyme regulates levels of major myofibrillar proteins in L6 muscle cells. Am. J. Physiol. Endocrinol. Metab. 2009, 297, 1283–1290. [Google Scholar] [CrossRef] [PubMed]
- Altun, M.; Zhao, B.; Velasco, K.; Liu, H.; Hassink, G.; Paschke, J.; Pereira, T.; Lindsten, K. Ubiquitin-specific protease 19 (USP19) regulates hypoxia-inducible factor 1α (HIF-1α) during hypoxia. J. Biol. Chem. 2012, 287, 1962–1969. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Hahn, A.A.; Hu, S.; Yang, X. The USP19 deubiquitinase regulates the stability of c-IAP1 and c-IAP2. J. Biol. Chem. 2011, 286, 35380–35387. [Google Scholar] [CrossRef] [PubMed]
- Wiles, B.; Miao, M.; Coyne, E.; Larose, L.; Cybulsky, A.V.; Wing, S.S. USP19 deubiquitinating enzyme inhibits muscle cell differentiation by suppressing unfolded-protein response signaling. Mol. Biol. Cell 2015, 26, 913–923. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.G.; Kim, W.; Gygi, S.; Ye, Y. Characterization of the deubiquitinating activity of USP19 and its role in endoplasmic reticulum-associated degradation. J. Biol. Chem. 2014, 289, 3510–3517. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, N.; Harada, K.; Kato, M.; Hirose, S. Ubiquitin-specific protease 19 regulates the stability of the E3 ubiquitin ligase MARCH6. Exp. Cell Res. 2014, 328, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Neutzner, A.; Neutzner, M.; Benischke, A.S.; Ryu, S.W.; Frank, S.; Youle, R.J.; Karbowski, M. A systematic search for endoplasmic reticulum (ER) membrane-associated ring finger proteins identifies Nixin/ZNRF4 as a regulator of calnexin stability and ER homeostasis. J. Biol. Chem. 2011, 286, 8633–8643. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Kato, M.; Nakamura, N.; Department of Life Science and Technology, Tokyo Institute of Technology, Yokohama, Japan. Unpublished data. 2016.
- Kikkert, M.; Doolman, R.; Dai, M.; Avner, R.; Hassink, G.; van Voorden, S.; Thanedar, S.; Roitelman, J.; Chau, V.; Wiertz, E. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J. Biol. Chem. 2004, 279, 3525–3534. [Google Scholar] [CrossRef] [PubMed]
- Christianson, J.C.; Ye, Y. Cleaning up in the endoplasmic reticulum: Ubiquitin in charge. Nat. Struct. Mol. Biol. 2014, 21, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, M.; Ishiguro, M.; Niinuma, Y.; Uesugi, M.; Nomura, Y. Human HRD1 protects against ER stress-induced apoptosis through ER-associated degradation. FEBS Lett. 2002, 532, 147–152. [Google Scholar] [CrossRef]
- Kaneko, M.; Nomura, Y. ER signaling in unfolded protein response. Life Sci. 2003, 74, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Nadav, E.; Shmueli, A.; Barr, H.; Gonen, H.; Ciechanover, A.; Reiss, Y. A novel mammalian endoplasmic reticulum ubiquitin ligase homologous to the yeast HRD1. Biochem. Biophys. Res. Commun. 2003, 303, 91–97. [Google Scholar] [CrossRef]
- Bernasconi, R.; Molinari, M. ERAD and ERAD tuning: Disposal of cargo and of ERAD regulators from the mammalian ER. Curr. Opin. Cell Biol. 2011, 23, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Blount, J.R.; Burr, A.A.; Denuc, A.; Marfany, G.; Todi, S.V. Ubiquitin-specific protease 25 functions in endoplasmic reticulum-associated degradation. PLoS ONE 2012, 7, e36542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, E.S.; Hong, H.; Kim, C.; Mook-Jung, I. Acute ER stress regulates amyloid precursor protein processing through ubiquitin-dependent degradation. Sci. Rep. 2015, 5, 8805–8814. [Google Scholar] [CrossRef] [PubMed]
- Husnjak, K.; Elsasser, S.; Zhang, N.; Chen, X.; Randles, L.; Shi, Y.; Hofmann, K.; Walters, K.J.; Finley, D.; Dikic, I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 2008, 453, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Schreiner, P.; Chen, X.; Husnjak, K.; Randles, L.; Zhang, N.; Elsasser, S.; Finley, D.; Dikic, I.; Walters, K.J.; Groll, M. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 2008, 453, 548–552. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Song, B.L. Ubiquitin ligases in cholesterol metabolism. Diabetes Metab. J. 2014, 38, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Baldridge, R.D.; Rapoport, T.A. Autoubiquitination of the Hrd1 ligase triggers protein retrotranslocation in ERAD. Cell 2016, 166, 394–407. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.G.; Takahama, S.; Zhang, G.; Tomarev, S.I.; Ye, Y. Unconventional secretion of misfolded proteins promotes adaptation to proteasome dysfunction in mammalian cells. Nat. Cell Biol. 2016, 18, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, N.; Fukuda, H.; Kato, A.; Hirose, S. MARCH-II is a syntaxin-6-binding protein involved in endosomal trafficking. Mol. Biol. Cell 2005, 16, 1696–1710. [Google Scholar] [CrossRef] [PubMed]



© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harada, K.; Kato, M.; Nakamura, N. USP19-Mediated Deubiquitination Facilitates the Stabilization of HRD1 Ubiquitin Ligase. Int. J. Mol. Sci. 2016, 17, 1829. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17111829
Harada K, Kato M, Nakamura N. USP19-Mediated Deubiquitination Facilitates the Stabilization of HRD1 Ubiquitin Ligase. International Journal of Molecular Sciences. 2016; 17(11):1829. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17111829
Chicago/Turabian StyleHarada, Kumi, Masako Kato, and Nobuhiro Nakamura. 2016. "USP19-Mediated Deubiquitination Facilitates the Stabilization of HRD1 Ubiquitin Ligase" International Journal of Molecular Sciences 17, no. 11: 1829. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17111829