Comparative Analysis of Toxic Responses of Organic Extracts from Diesel and Selected Alternative Fuels Engine Emissions in Human Lung BEAS-2B Cells
Abstract
:1. Introduction
2. Results
2.1. Characteristics of Collected Exhaust Particles; Chemical Analysis of Organic Extracts
2.2. Cytotoxicity Assessment
2.3. Quantification of Intracellularly Generated Reactive Oxygen Species and Reduced Glutathione Level
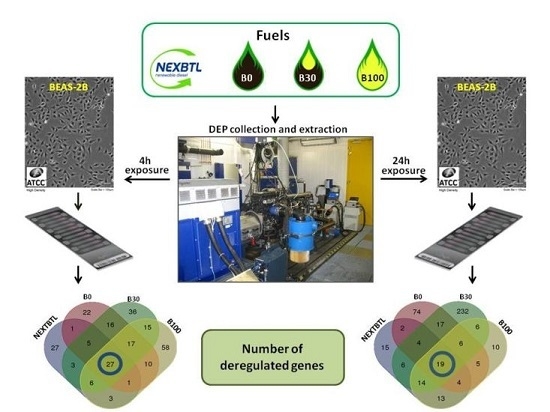
2.4. Gene Expression Profiling in BEAS-2B Exposed to Diverse DEP Extracts
2.4.1. Identification of Differentially Expressed Genes
2.4.2. Functional Analysis
Commonly Modulated Pathways
Analysis of Variance
2.4.3. Quantitative RT-PCR Validation of Selected Genes
3. Discussion
3.1. Common Cellular Response—4 h Cell Exposure
3.2. Differential Response Detected by Analysis of Variance—4 h Cell Exposure
3.3. Common Cellular Response—24 h Cell Exposure
3.4. Differential Response Detected by Analysis of Variance—24 h Cell Exposure
3.5. Study Limitations
4. Materials and Methods
4.1. Chemicals and Biochemicals
4.2. Test Vehicle and Exhaust Particles Collection
4.3. Particle Characterization and Chemical Analysis
4.4. Cell Cultures and Exposure Conditions
4.5. Cytotoxicity
4.6. Cell Culture Conditions, RNA Isolation and Quality Control
4.7. Microarray Analysis
4.8. Real-Time Quantitative PCR (RT-qPCR) Verification
4.9. Quantification of Intracellular ROS
4.10. Quantification of Glutathione Levels
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Aatola, H.; Larmi, M.; Sarjovaara, T.; Mikkonen, S. Hydrotreated vegetable oil (HVO) as a renewable diesel fuel: Trade-off between nox, particulate emission, and fuel consumption of a heavy duty engine. SAE Int. J. Engines 2009, 1, 1251–1262. [Google Scholar] [CrossRef]
- Moser, B.R. Impact of fatty ester composition on low temperature properties of biodiesel–petroleum diesel blends. Fuel 2014, 115, 500–506. [Google Scholar] [CrossRef]
- Kim, D.; Kim, S.; Oh, S.; No, S.-Y. Engine performance and emission characteristics of hydrotreated vegetable oil in light duty diesel engines. Fuel 2014, 125, 36–43. [Google Scholar] [CrossRef]
- Omidvarborna, H.; Kumar, A.; Kim, D.-S. Variation of diesel soot characteristics by different types and blends of biodiesel in a laboratory combustion chamber. Sci. Total Environ. 2016, 544, 450–459. [Google Scholar] [CrossRef] [PubMed]
- Prokopowicz, A.; Zaciera, M.; Sobczak, A.; Bielaczyc, P.; Woodburn, J. The effects of neat biodiesel and biodiesel and HVO blends in diesel fuel on exhaust emissions from a light duty vehicle with a diesel engine. Environ. Sci. Technol. 2015, 49, 7473–7482. [Google Scholar] [CrossRef] [PubMed]
- Rakopoulos, D.C.; Rakopoulos, C.D.; Giakoumis, E.G. Impact of properties of vegetable oil, bio-diesel, ethanol and n-butanol on the combustion and emissions of turbocharged hddi diesel engine operating under steady and transient conditions. Fuel 2015, 156, 1–19. [Google Scholar] [CrossRef]
- Singh, D.; Subramanian, K.A.; Singal, S.K. Emissions and fuel consumption characteristics of a heavy duty diesel engine fueled with hydroprocessed renewable diesel and biodiesel. Appl. Energy 2015, 155, 440–446. [Google Scholar] [CrossRef]
- Woo, C.; Kook, S.; Hawkes, E.R.; Rogers, P.L.; Marquis, C. Dependency of engine combustion on blending ratio variations of lipase-catalysed coconut oil biodiesel and petroleum diesel. Fuel 2016, 169, 146–157. [Google Scholar] [CrossRef]
- Xue, J.; Grift, T.E.; Hansen, A.C. Effect of biodiesel on engine performances and emissions. Renew. Sustain. Energy Rev. 2011, 15, 1098–1116. [Google Scholar] [CrossRef]
- Millo, F.; Debnath, B.K.; Vlachos, T.; Ciaravino, C.; Postrioti, L.; Buitoni, G. Effects of different biofuels blends on performance and emissions of an automotive diesel engine. Fuel 2015, 159, 614–627. [Google Scholar] [CrossRef]
- Lapuerta, M.; Armas, O.; Rodríguez-Fernández, J. Effect of biodiesel fuels on diesel engine emissions. Prog. Energy Combust. Sci. 2008, 34, 198–223. [Google Scholar] [CrossRef]
- Vojtisek-Lom, M.; Pechout, M.; Dittrich, L.; Beranek, V.; Kotek, M.; Schwarz, J.; Vodicka, P.; Milcova, A.; Rossnerova, A.; Ambroz, A.; et al. Polycyclic aromatic hydrocarbons (PAH) and their genotoxicity in exhaust emissions from a diesel engine during extended low-load operation on diesel and biodiesel fuels. Atmos. Environ. 2015, 109, 9–18. [Google Scholar] [CrossRef]
- Tang, S.; LaDuke, G.; Chien, W.; Frank, B.P. Impacts of biodiesel blends on pm2.5, particle number and size distribution, and elemental/organic carbon from nonroad diesel generators. Fuel 2016, 172, 11–19. [Google Scholar] [CrossRef]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Chemical agents and related occupations. IARC Monogr. Eval. Carcinog. Risks Hum. 2012, 100, 9–562. [Google Scholar]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Diesel and gasoline engine exhausts and some nitroarenes. IARC monographs on the evaluation of carcinogenic risks to humans. IARC Monogr. Eval. Carcinog. Risks Hum. 2014, 105, 9–699. [Google Scholar]
- Claxton, L.D. The history, genotoxicity and carcinogenicity of carbon-based fuels and their emissions: Part 4—Alternative fuels. Mutat. Res. Rev. Mutat. Res. 2015, 763, 86–102. [Google Scholar] [CrossRef] [PubMed]
- Schins, R.P.; Knaapen, A.M. Genotoxicity of poorly soluble particles. Inhal. Toxicol. 2007, 19, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Topinka, J.; Milcova, A.; Schmuczerova, J.; Mazac, M.; Pechout, M.; Vojtisek-Lom, M. Genotoxic potential of organic extracts from particle emissions of diesel and rapeseed oil powered engines. Toxicol. Lett. 2012, 212, 11–17. [Google Scholar] [CrossRef] [PubMed]
- André, V.; Barraud, C.; Capron, D.; Preterre, D.; Keravec, V.; Vendeville, C.; Cazier, F.; Pottier, D.; Morin, J.P.; Sichel, F. Comparative mutagenicity and genotoxicity of particles and aerosols emitted by the combustion of standard vs. Rapeseed methyl ester supplemented bio-diesel fuels: Impact of after treatment devices: Oxidation catalyst and particulate filter. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2015, 777, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Steiner, S.; Heeb, N.V.; Czerwinski, J.; Comte, P.; Mayer, A.; Petri-Fink, A.; Rothen-Rutishauser, B. Test-methods on the test-bench: A comparison of complete exhaust and exhaust particle extracts for genotoxicity/mutagenicity assessment. Environ. Sci. Technol. 2014, 48, 5237–5244. [Google Scholar] [CrossRef] [PubMed]
- Iba, M.M.; Caccavale, R.J. Genotoxic bioactivation of constituents of a diesel exhaust particle extract by the human lung. Environ. Mol. Mutagen. 2013, 54, 158–171. [Google Scholar] [CrossRef] [PubMed]
- Bao, L.; Xu, A.; Tong, L.; Chen, S.; Zhu, L.; Zhao, Y.; Zhao, G.; Jiang, E.; Wang, J.; Wu, L. Activated toxicity of diesel particulate extract by ultraviolet a radiation in mammalian cells: Role of singlet oxygen. Environ. Health Perspect. 2009, 117, 436–441. [Google Scholar] [CrossRef] [PubMed]
- Jalava, P.I.; Tapanainen, M.; Kuuspalo, K.; Markkanen, A.; Hakulinen, P.; Happo, M.S.; Pennanen, A.S.; Ihalainen, M.; Yli-Pirila, P.; Makkonen, U.; et al. Toxicological effects of emission particles from fossil- and biodiesel-fueled diesel engine with and without doc/poc catalytic converter. Inhal. Toxicol. 2010, 22, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Kooter, I.M.; van Vugt, M.A.T.M.; Jedynska, A.D.; Tromp, P.C.; Houtzager, M.M.G.; Verbeek, R.P.; Kadijk, G.; Mulderij, M.; Krul, C.A.M. Toxicological characterization of diesel engine emissions using biodiesel and a closed soot filter. Atmos. Environ. 2011, 45, 1574–1580. [Google Scholar] [CrossRef]
- Palkova, L.; Vondracek, J.; Trilecova, L.; Ciganek, M.; Pencikova, K.; Neca, J.; Milcova, A.; Topinka, J.; Machala, M. The aryl hydrocarbon receptor-mediated and genotoxic effects of fractionated extract of standard reference diesel exhaust particle material in pulmonary, liver and prostate cells. Toxicol. In Vitro 2015, 29, 438–448. [Google Scholar] [CrossRef] [PubMed]
- Lou, H.; Du, S.Y.; Ji, Q.; Stolz, A. Induction of AKR1C2 by phase II inducers: Identification of a distal consensus antioxidant response element regulated by Nrf2. Mol. Pharmacol. 2006, 69, 1662–1672. [Google Scholar] [CrossRef] [PubMed]
- Penning, T.M. Human aldo-keto reductases and the metabolic activation of polycyclic aromatic hydrocarbons. Chem. Res. Toxicol. 2014, 27, 1901–1917. [Google Scholar] [CrossRef] [PubMed]
- Longhin, E.; Capasso, L.; Battaglia, C.; Proverbio, M.C.; Cosentino, C.; Cifola, I.; Mangano, E.; Camatini, M.; Gualtieri, M. Integrative transcriptomic and protein analysis of human bronchial BEAS-2B exposed to seasonal urban particulate matter. Environ. Pollut. 2016, 209, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q. Role of Nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401–426. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wang, M.Y.; Oberley, T.D.; Sempf, J.M.; Nel, A.E. Comparison of the pro-oxidative and proinflammatory effects of organic diesel exhaust particle chemicals in bronchial epithelial cells and macrophages. J. Immunol. 2002, 169, 4531–4541. [Google Scholar] [CrossRef] [PubMed]
- Totlandsdal, A.I.; Lag, M.; Lilleaas, E.; Cassee, F.; Schwarze, P. Differential proinflammatory responses induced by diesel exhaust particles with contrasting pah and metal content. Environ. Toxicol. 2015, 30, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Arrigo, A.P. The cellular “networking” of mammalian hsp27 and its functions in the control of protein folding, redox state and apoptosis. Adv. Exp. Med. Biol. 2007, 594, 14–26. [Google Scholar] [PubMed]
- Krause, K.; Wasner, M.; Reinhard, W.; Haugwitz, U.; Dohna, C.L.Z.; Mossner, J.; Engeland, K. The tumour suppressor protein p53 can repress transcription of cyclin B. Nucleic Acids Res. 2000, 28, 4410–4418. [Google Scholar] [CrossRef] [PubMed]
- Karantanos, T.; Tanimoto, R.; Edamura, K.; Hirayama, T.; Yang, G.; Golstov, A.A.; Wang, J.X.; Kurosaka, S.; Park, S.; Thompson, T.C. Systemic GLIPR1-ΔTM protein as a novel therapeutic approach for prostate cancer. Int. J. Cancer 2014, 134, 2003–2013. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Liu, X.; Zhang, W.; Xiao, W.H. P53 directly suppresses BNIP3 expression to protect against hypoxia-induced cell death. EMBO J. 2011, 30, 3397–3415. [Google Scholar] [CrossRef] [PubMed]
- Galvagni, F.; Orlandini, M.; Oliviero, S. Role of the AP-1 transcription factor FOSL1 in endothelial cell adhesion and migration. Cell Adhes. Migr. 2013, 7, 408–411. [Google Scholar] [CrossRef] [PubMed]
- West, M.D.; Shay, J.W.; Wright, W.E.; Linskens, M.H.K. Altered expression of plasminogen activator and plasminogen activator inhibitor during cellular senescence. Exp. Gerontol. 1996, 31, 175–193. [Google Scholar] [CrossRef]
- Johannson, K.A.; Vittinghoff, E.; Lee, K.; Balmes, J.R.; Ji, W.; Kaplan, G.G.; Kim, D.S.; Collard, H.R. Acute exacerbation of idiopathic pulmonary fibrosis associated with air pollution exposure. Eur. Respir. J. 2014, 43, 1124–1131. [Google Scholar] [CrossRef] [PubMed]
- Hattori, N.; Mizuno, S.; Yoshida, Y.; Chin, K.; Mishima, M.; Sisson, T.H.; Simon, R.H.; Nakamura, T.; Miyake, M. The plasminogen activation system reduces fibrosis in the lung by a hepatocyte growth factor-dependent mechanism. Am. J. Pathol. 2004, 164, 1091–1098. [Google Scholar] [CrossRef]
- Cho, H.Y.; Reddy, S.P.; Yamamoto, M.; Kleeberger, S.R. The transcription factor Nrf2 protects against pulmonary fibrosis. FASEB J. 2004, 18, 1258–1260. [Google Scholar] [CrossRef] [PubMed]
- Lakatos, H.F.; Thatcher, T.H.; Kottmann, R.M.; Garcia, T.M.; Phipps, R.P.; Sime, P.J. The role of ppars in lung fibrosis. PPAR Res. 2007, 2007, 71323. [Google Scholar] [CrossRef] [PubMed]
- Devchand, P.R.; Ziouzenkova, O.; Plutzky, J. Oxidative stress and peroxisome proliferator-activated receptors: Reversing the curse? Circ. Res. 2004, 95, 1137–1139. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jin, Y.; Huang, M.; Penning, T.M. The role of human Aldo-Keto reductases in the metabolic activation and detoxication of polycyclic aromatic hydrocarbons: Interconversion of pah catechols and pah o-quinones. Front. Pharmacol. 2012, 3, 193. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Vudathala, D.K.; Blair, I.A.; Penning, T.M. Competing roles of Aldo-Keto reductase 1A1 and cytochrome P4501B1 in benzo[a]pyrene-7,8-diol activation in human bronchoalveolar H358 cells: Role of akrs in p4501b1 induction. Chem. Res. Toxicol. 2006, 19, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Andrysik, Z.; Vondracek, J.; Marvanova, S.; Ciganek, M.; Neca, J.; Pencikova, K.; Mahadevan, B.; Topinka, J.; Baird, W.M.; Kozubik, A.; et al. Activation of the aryl hydrocarbon receptor is the major toxic mode of action of an organic extract of a reference urban dust particulate matter mixture: The role of polycyclic aromatic hydrocarbons. Mutat. Res. Fund. Mol. M 2011, 714, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Crane, R.; Gadea, B.; Littlepage, L.; Wu, H.; Ruderman, J.V. Aurora A, meiosis and mitosis. Biol. Cell 2004, 96, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Kunitoku, N.; Sasayama, T.; Marumoto, T.; Zhang, D.W.; Honda, S.; Kobayashi, O.; Hatakeyama, K.; Ushio, Y.; Saya, H.; Hirota, T. Cenp-a phosphorylation by aurora-A in prophase is required for enrichment of aurora-b inner centromeres and for kinetochore function. Dev. Cell 2003, 5, 853–864. [Google Scholar] [CrossRef]
- Neef, R.; Preisinger, C.; Sutcliffe, J.; Kopajtich, R.; Nigg, E.A.; Mayer, T.U.; Barr, F.A. Phosphorylation of mitotic kinesin-like protein 2 by polo-like kinase 1 is required for cytokinesis. J. Cell Biol. 2003, 162, 863–875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.C.; Yang, T.Y.; Yu, C.T.; Phan, L.; Ivan, C.; Sood, A.K.; Hsu, S.L.; Lee, M.H. P53 negatively regulates aurora a via both transcriptional and posttranslational regulation. Cell Cycle 2012, 11, 3433–3442. [Google Scholar] [CrossRef] [PubMed]
- Eyers, P.A.; Erikson, E.; Chen, L.G.; Maller, J.L. A novel mechanism for activation of the protein kinase aurora A. Curr. Biol. 2003, 13, 691–697. [Google Scholar] [CrossRef]
- Garrido, G.; Vernos, I. Non-centrosomal tpx2-dependent regulation of the aurora a kinase: Functional implications for healthy and pathological cell division. Front. Oncol. 2016, 6, 88. [Google Scholar] [CrossRef] [PubMed]
- Aguirre-Portoles, C.; Bird, A.W.; Hyman, A.; Canamero, M.; de Castro, I.P.; Malunnbres, M. Tpx2 controls spindle integrity, genome stability, and tumor development. Cancer Res. 2012, 72, 1518–1528. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Kibbe, W.A.; Lin, S.M. Lumi: A pipeline for processing illumina microarray. Bioinformatics 2008, 24, 1547–1548. [Google Scholar] [CrossRef] [PubMed]
- Smyth, G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3, Article 3. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. Toppgene suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R foundation for Statistical Computing: Vienna, Austria, 2015. Available online: http://www.R-project.org/ (accessed on 8 August 2016).
- Chambers, J.M.; Freeny, A.; Heiberger, R.M. Analysis of Variance; Designed Experiments; Wadsworth & Brooks/Cole: Pacific Grove, CA, USA, 1992. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar]
- Yandell, B.S. Practical Data Analysis for Designed Experiments; Crc Press, Chapman & Hall: London, UK, 1997; Volume 39. [Google Scholar]









| PAHs | ||||
| Compound (ng/mg DEP) | B0 | B30 | B100 | NEXBTL100 |
| Fluoranthene | 41.9 | 110 | 117 | 39.4 |
| Pyrene | 67.7 | 152 | 144 | 43.1 |
| Benz[a]anthracene * | 6.6 | 9.8 | 26.1 | 5.0 |
| Chrysene * | 5.1 | 6.0 | 12.5 | 1.0 |
| Benzo[b]fluoranthene * | 1.2 | 1.4 | 7.0 | 0.3 |
| Benzo[k]fluoranthene * | n.d. | n.d. | 2.6 | n.d. |
| Benzo[a]pyrene * | 0.6 | 0.9 | 1.7 | 1.2 |
| Benzo[g,h,i]perylene | 2.2 | 2.8 | 3.8 | 2.0 |
| Dibenz[a,h]anthracene * | n.d. | n.d. | n.d. | n.d. |
| Indeno[1,2,3-cd]pyrene * | 0.7 | 0.8 | 1.8 | n.d. |
| 4H-Cyclopenta[d,e,f]phenathrene | 2.8 | 3.8 | 4.3 | 9.4 |
| Benzo[c]phenanthrene | 1.9 | 3.8 | 4.0 | 1.3 |
| Benzo[j]fluoranthene | 2.4 | 2.8 | 6.4 | 2.0 |
| Benzo[e]pyrene | 0.7 | 0.6 | 3.6 | 0.3 |
| Triphenylene | 9.0 | 3.6 | 4.0 | 1.1 |
| Benzo[c]chrysene | n.d. | n.d. | 0.2 | n.d. |
| Coronene | 0.8 | 0.7 | 0.7 | 0.6 |
| Methylated PAHs | ||||
| Compound (ng/mg DEP) | B0 | B30 | B100 | NEXBTL100 |
| 1-Methylpyrene | 5.1 | 7.2 | 3.9 | 1.4 |
| 2-Methylpyrene | 8.5 | 11.0 | 8.0 | 2.9 |
| 4-Methylpyrene | 10.0 | 13.4 | 12 | 3.7 |
| 1-Methylchrysene | 0.3 | 0.3 | 1.2 | n.d. |
| 7-Methylbenz[a]anthracene | 0.3 | n.d. | n.d. | 0.2 |
| Oxygenated PAHs | ||||
| Compound (ng/mg DEP) | B0 | B30 | B100 | NEXBTL100 |
| 1,8-Naphthalic Anhydride | 25.4 | 29.5 | 29.7 | 19.8 |
| Phenanthrene-9,10-dione | 1.3 | 0.8 | 0.8 | 0.5 |
| 9H-Fluoren-9-one | 4.3 | 2.8 | 1.7 | 4.7 |
| Anthrone | n.d. | n.d. | n.d. | n.d. |
| Anthracene-9,10-dione | 2.7 | 4.5 | 4.0 | 2.6 |
| 7H-Benz[d,e]anthracene-7-one | 1.4 | 1.5 | 2.4 | 0.28 |
| 9-Hydroxybenzo[a]pyrene | 0.06 | 0.07 | 0.09 | 0.07 |
| Benz[a]anthracene-7,12-dione | 0.02 | 0.04 | 0.15 | 0.01 |
| 3-Hydroxybenzo[a]pyrene | 0.05 | 0.05 | 0.10 | 0.02 |
| Nitrated PAHs | ||||
| Compound (pg/mg DEP) | B0 | B30 | B100 | NEXBTL100 |
| 1-Nitropyrene | 314 | 539 | 1504 | 126 |
| 2-Nitropyrene | 25.0 | 12.0 | 23.0 | n.d. |
| 4-Nitropyrene | 18.0 | 16.0 | 44.0 | 10.0 |
| 3-Nitrofluoranthene | 7.0 | 16.0 | 41.0 | 2.0 |
| Dinitrated PAHs | ||||
| Compound (pg/mg DEP) | B0 | B30 | B100 | NEXBTL100 |
| 1,3-Dinitropyrene | 1.6 | 2.4 | 17.7 | 0.4 |
| 1,6-Dinitropyrene | 1.2 | 5.6 | 65.0 | n.d. |
| 1,8-Dinitropyrene | 0.9 | 4.6 | 48.0 | n.d. |
| Name | Genes from Input |
|---|---|
| ”Benzo[a]pyrene metabolism”, ”Synthesis of bile acids and bile salts via 27-hydroxycholesterol”, ”Synthesis of bile acids and bile salts via 24-hydroxycholesterol”, ”Synthesis of bile acids and bile salts via 7α-hydroxycholesterol”, ”Synthesis of bile acids and bile salts”, ”Bile acid and bile salt metabolism” | ↑ AKR1C3, AKR1C2 |
| ”Metabolism of lipids and lipoproteins” | ↑ TXNRD1, GLIPR1, AKR1C3, AKR1C2; ↓ LDLR, CTGF |
| ”PPARA Activates Gene Expression”, ”Regulation of Lipid Metabolism by Peroxisome proliferator-activated receptor alpha (PPARalpha) “ | ↑ TXNRD1, GLIPR1; ↓ CTGF |
| ”Retinoid metabolism and transport, Ovarian steroidogenesis” | ↑ AKR1C3; ↓ LDLR |
| ”Oxidative Stress” | ↑ TXNRD1, HMOX1 |
| ”Keap1-Nrf2 Pathway” | ↑ HMOX1, GCLM |
| ”Validated transcriptional targets of AP1 family members Fra1 and Fra2” | ↑ PLAU, HMOX1 |
| ”heme catabolic” | ↑ HMOX1 |
| ”Dissolution of Fibrin Clot”, Fibrinolysis Pathway”, ”Plasminogen activating cascade”, ”Blood Clotting Cascade”, ”Blood coagulation” | ↑ PLAU, SERPINB2 |
| ”Senescence and Autophagy” | ↑ IL24, PLAU, SERPINB2 |
| ”amb2 Integrin signaling” | ↑ PLAU; ↓ CTGF |
| ”DNA damage response (only ATM dependent) “ | ↑ PLAU, BIK; ↓ LDLR |
| ”intrinsic apoptotic” | ↑ BIK; ↓ BNIP3 |
| ”t(4;14) translocations of FGFR3” | ↑ FGFR3 |
| “Bladder cancer” | ↑ FGFR3; ↓ E2F2 |
| ”MicroRNAs in cancer” | ↑ PLAU, FGFR3, HMOX1; ↓ E2F2 |
| ”p38 signaling mediated by MAPKAP kinases” | ↑ HSPB1, SFN |
| Name | Genes from Input |
|---|---|
| ”Benzo[a]pyrene metabolism”, ”Synthesis of bile acids and bile salts via 27-hydroxycholesterol”, ”Synthesis of bile acids and bile salts via 24-hydroxycholesterol”, ”Synthesis of bile acids and bile salts via 7α-hydroxycholesterol”, ”Synthesis of bile acids and bile salts”, ”Bile acid and bile salt metabolism”, ”Steroid hormone biosynthesis”, ”Metabolism of xenobiotics by cytochrome P450” | ↑ AKR1C4, AKR1C2 |
| ”Metabolism of lipids and lipoproteins” | ↑ TXNRD1, AKR1C4, AKR1C2; ↓ HMGCS1 |
| ”ketone bodies metabolic” | ↓ HMGCS1 |
| ”Selenium Pathway” | ↑ TXNRD1, IL1B, KYNU |
| ”thioredoxin pathway” | ↑ TXNRD1 |
| ”Genes encoding collagen proteins”, ”Assembly of collagen fibrils and other multimeric structures”, ”Collagen biosynthesis and modifying enzymes”, ”Collagen formation” | ↑ COL7A1; ↓ COL8A1 |
| ”Protein digestion and absorption” | ↑ COL7A1, SLC3A2 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Libalova, H.; Rossner,, P., Jr.; Vrbova, K.; Brzicova, T.; Sikorova, J.; Vojtisek-Lom, M.; Beranek, V.; Klema, J.; Ciganek, M.; Neca, J.; et al. Comparative Analysis of Toxic Responses of Organic Extracts from Diesel and Selected Alternative Fuels Engine Emissions in Human Lung BEAS-2B Cells. Int. J. Mol. Sci. 2016, 17, 1833. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17111833
Libalova H, Rossner, P Jr., Vrbova K, Brzicova T, Sikorova J, Vojtisek-Lom M, Beranek V, Klema J, Ciganek M, Neca J, et al. Comparative Analysis of Toxic Responses of Organic Extracts from Diesel and Selected Alternative Fuels Engine Emissions in Human Lung BEAS-2B Cells. International Journal of Molecular Sciences. 2016; 17(11):1833. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17111833
Chicago/Turabian StyleLibalova, Helena, Pavel Rossner,, Jr., Kristyna Vrbova, Tana Brzicova, Jitka Sikorova, Michal Vojtisek-Lom, Vit Beranek, Jiri Klema, Miroslav Ciganek, Jiri Neca, and et al. 2016. "Comparative Analysis of Toxic Responses of Organic Extracts from Diesel and Selected Alternative Fuels Engine Emissions in Human Lung BEAS-2B Cells" International Journal of Molecular Sciences 17, no. 11: 1833. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17111833