Generation and Characterisation of a Reference Transcriptome for Lentil (Lens culinaris Medik.)
Abstract
:1. Introduction
2. Results
2.1. De Novo Sequence Assembly
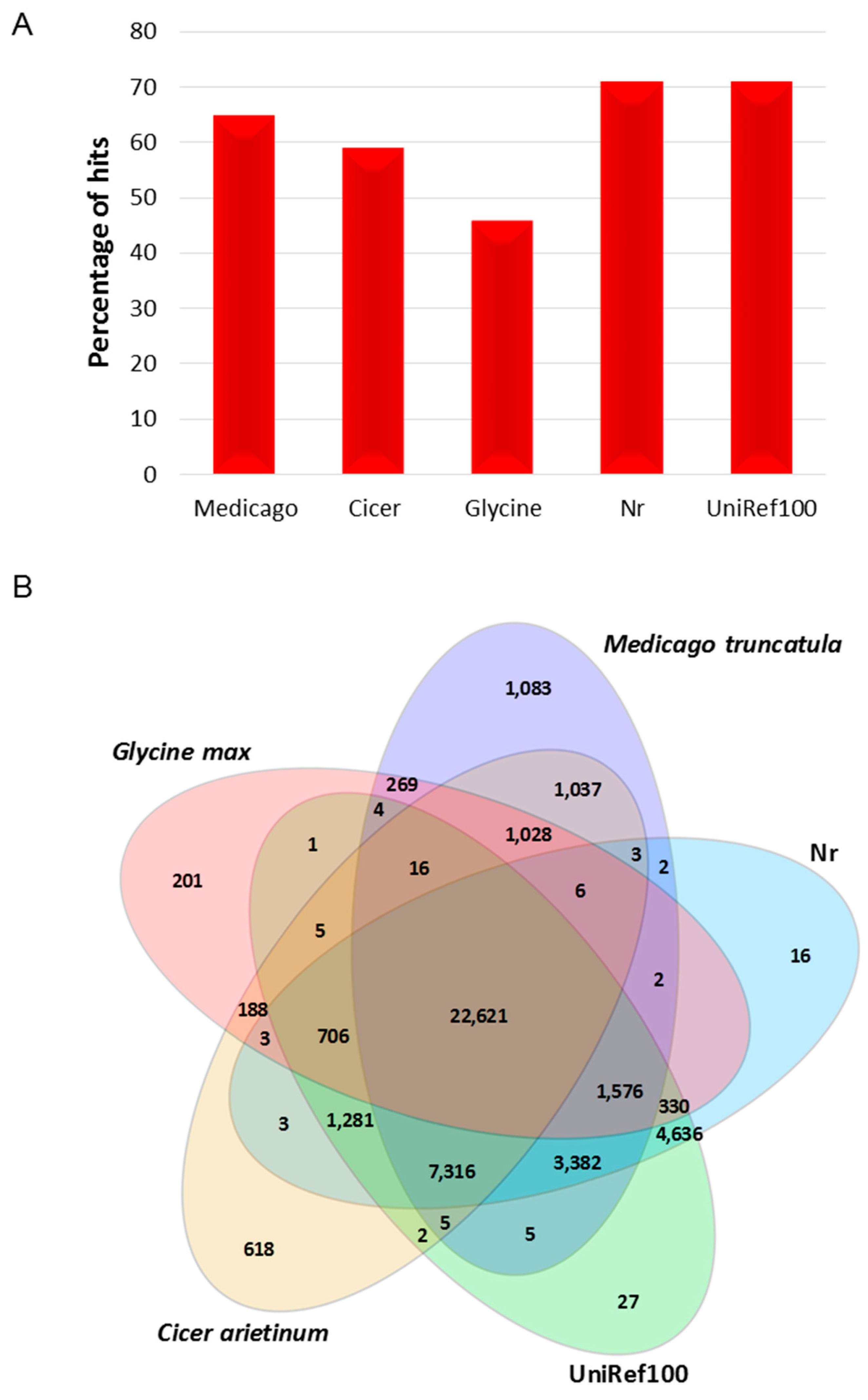
2.2. Functional Annotation and Classification
2.3. Tissue-Specific Expression Analysis
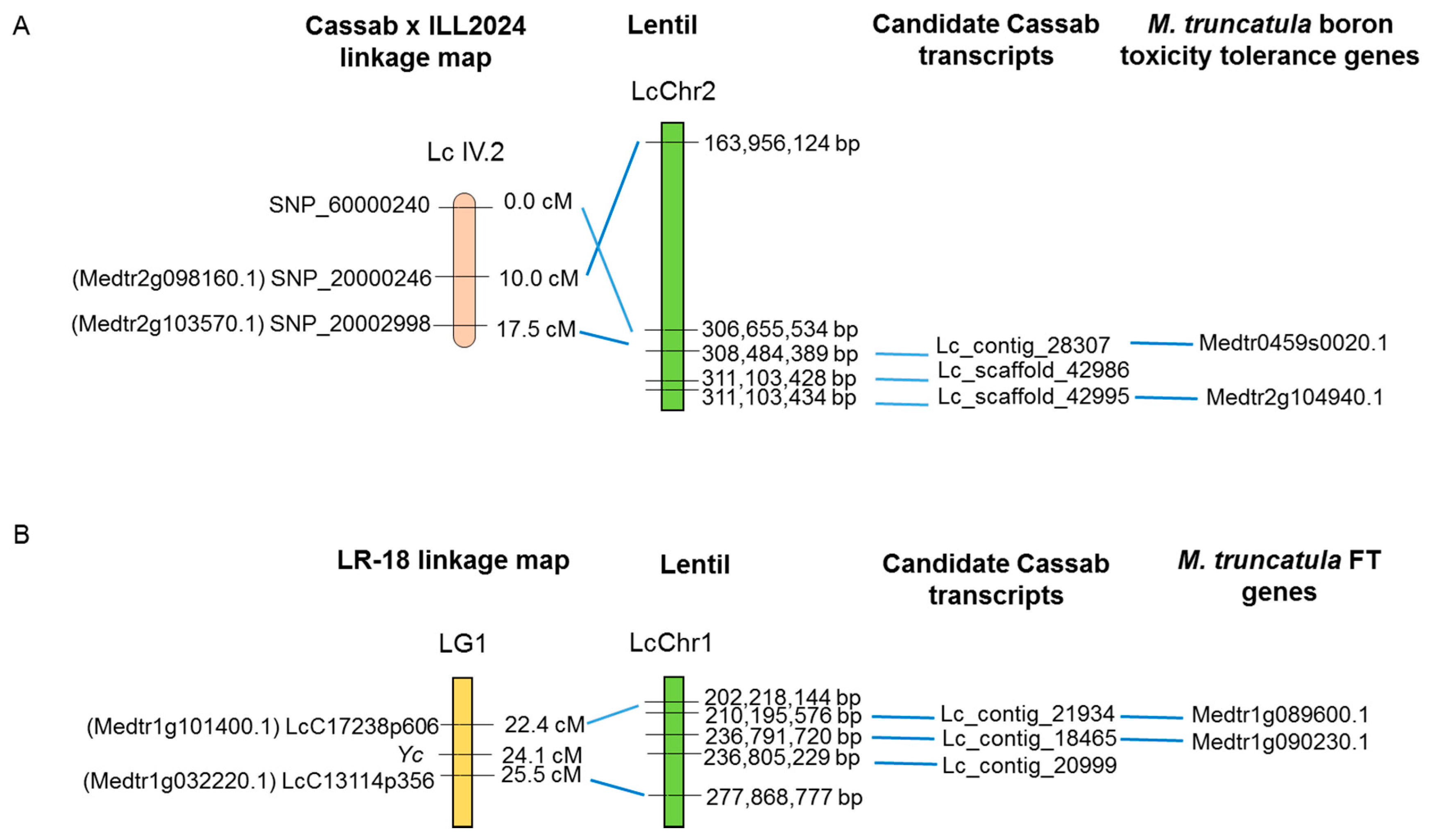
2.4. Identification of Candidate Genes for Tolerance to Boron Toxicity and Flowering Time in the Reference Transcriptome
3. Discussion
3.1. Reference Transcriptome Assembly Characteristics
3.2. Annotation of Gene Sequences
3.3. Assessment of Tissue-Specific Gene Expression
3.4. Identification of Candidate Genes for Tolerance to B Toxicity and Flowering Time in the Reference Transcriptome
4. Experimental Section
4.1. Plant Materials
4.2. Library Preparation
4.3. De Novo Transcriptome Sequence Assembly
4.4. Functional Annotation and Classification of Reference Transcriptome
4.5. Tissue-Specific Expression Analysis
4.6. Identification of Candidate Genes for Tolerance to B Toxicity and Flowering Time in the Reference Transcriptome
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| B | Boron |
| bp | Base pair |
| BLAST | Basic Local Alignment Search Tool |
| cDNA | Complementary DNA |
| CDS | Coding DNA sequences |
| CO | CONSTANS |
| DNA | Deoxyribonucleic acid |
| EST | Expressed sequence tag |
| FT | Flowering time |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| Gbp | Giga base pair |
| GBS | Genotyping-by-sequencing |
| GO | Gene Ontology |
| HC | High confidence |
| KAAS | KEGG Automatic Annotation Server |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LOD | Logarithm (base 10) of odds |
| Mb | Mega base pair |
| MIP | Major intrinsic protein |
| mRNA | Messenger RNA |
| NCBI | National Center for Biotechnology Information |
| NGS | Next-generation sequencing |
| Nr | Non-redundant |
| ORF | Open reading frame |
| PCA | Principal component analysis |
| qRT-PCR | Quantitative reverse transcription polymerase chain reaction |
| RNA | Ribonucleic acid |
| RNA-Seq | RNA sequencing technology |
| UTRs | Untranslated regions |
| W/V | Weight/volume |
References
- Garg, R.; Jai, M. RNA-Seq for transcriptome analysis in non-model plants. Methods Mol. Biol. 2013, 1069, 43–58. [Google Scholar] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chiodini, R.; Badr, A.; Zhang, Z. The impact of next-generation sequencing on genomics. J. Genet. Genom. 2011, 38, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Sudheesh, S.; Sawbridge, T.; Cogan, N.O.I.; Kennedy, P.; Forster, J.W.; Kaur, S. De novo assembly and characterisation of the field pea transcriptome using RNA-Seq. BMC Genom. 2015, 16, 611. [Google Scholar] [CrossRef] [PubMed]
- Cubero, J.I.; de la Perez, V.M.; Fratini, R. Origin, phylogeny, domestication and spread. In The Lentil: Botany, Production and Uses; Erskine, W., Muehlbauer, F.J., Sarker, A., Sharma, B., Eds.; CABI: Wallingford, UK, 2009; pp. 13–33. [Google Scholar]
- Nene, Y. Indian pulses through the millennia. Asian Agric. Hist. 2006, 10, 179–202. [Google Scholar]
- Yadav, S.S.; McNeil, D.L.; Stevenson, P.C. Lentil: An Ancient Crop for Modern Times; Springer: Dordrecht, The Netherlands, 2007. [Google Scholar]
- Sharpe, A.G.; Ramsay, L.; Sanderson, L.A.; Fedoruk, M.J.; Clarke, W.E.; Rong, L.; Kagale, S.; Vijayan, P.; Vandenberg, A.; Bett, K.E. Ancient orphan crop joins modern era: Gene-based SNP discovery and mapping in lentil. BMC Genom. 2013, 14, 192. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.K.; Barpete, S.; Kumar, J.; Gupta, P.; Sarker, A. Global lentil production: Constraints and strategies. SATSA Mukhapatra–Annu. Tech. Issue 2013, 17, 1–13. [Google Scholar]
- Kumar, S.; Rajendran, K.; Kumar, J.; Hamweih, A.; Baum, M. Current knowledge in lentil genomics and its application for crop improvement. Front. Plant Sci. 2015, 6, 78. [Google Scholar] [CrossRef] [PubMed]
- Arumuganathan, K.; Earle, E. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 1991, 9, 208–218. [Google Scholar] [CrossRef]
- Bett, K.; Ramsay, L.; Chan, C.; Sharpe, A.; Cook, D.; Penmetsa, R.V.; Chang, P.; Coyne, C.; McGee, R.; Main, D.; et al. Lentil v1.0 and beyond. In Proceedings of the XXIV Plant and Animal Genome Conference, San Diego, CA, USA, 9–13 January 2016.
- KnowPulse. Available online: http://knowpulse.usask.ca/portal/ (accessed on 23 May 2016).
- Zonneveld, B.; Leitch, I.; Bennett, M. First nuclear DNA amounts in more than 300 angiosperms. Ann. Bot. 2005, 96, 229–244. [Google Scholar] [CrossRef] [PubMed]
- Rabinowicz, P.D.; Schutz, K.; Dedhia, N.; Yordan, C.; Parnell, L.D.; Stein, L.; McCombie, W.R.; Martienssen, R.A. Differential methylation of genes and retrotransposons facilitates shotgun sequencing of the maize genome. Nat. Genet. 1999, 23, 305–308. [Google Scholar] [PubMed]
- Peterson, D.G.; Schulze, S.R.; Sciara, E.B.; Lee, S.A.; Bowers, J.E.; Nagel, A.; Jiang, N.; Tibbitts, D.C.; Wessler, S.R.; Paterson, A.H. Integration of Cot analysis, DNA cloning, and high-throughput sequencing facilitates genome characterization and gene discovery. Genome Res. 2002, 12, 797–807. [Google Scholar] [CrossRef]
- Kaur, S.; Cogan, N.O.I.; Pembleton, L.W.; Shinozuka, M.; Savin, K.W.; Materne, M.; Forster, J.W. Transcriptome sequencing of lentil based on second-generation technology permits large-scale unigene assembly and SSR marker discovery. BMC Genom. 2011, 12, 265. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Pembleton, L.; Cogan, N.O.I.; Savin, K.W.; Leonforte, T.; Paull, J.; Materne, M.; Forster, J.W. Transcriptome sequencing of field pea and faba bean for discovery and validation of SSR genetic markers. BMC Genom. 2012, 13, 104. [Google Scholar] [CrossRef] [PubMed]
- Mardis, E.R. The impact of next-generation sequencing echnology on genetics. Trends Genet. 2008, 24, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.J.; Dhingra, A.; Soltis, P.S.; Shaw, R.; Farmerie, W.G.; Folta, K.M.; Soltis, D.E. Rapid and accurate pyrosequencing of angiosperm plastid genomes. BMC Plant Biol. 2006, 6, 17. [Google Scholar] [CrossRef] [PubMed]
- Sierro, N.; Battey, J.N.; Ouadi, S.; Bovet, L.; Goepfert, S.; Bakaher, N.; Peitsch, M.C.; Ivanov, N.V. Reference genomes and transcriptomes of Nicotiana sylvestris and Nicotiana tomentosiformis. Genome Biol. 2013, 14, R60. [Google Scholar] [CrossRef] [PubMed]
- Alves-Carvalho, S.; Aubert, G.; Carrère, S.; Cruaud, C.; Brochot, A.; Jacquin, F.; Klein, A.; Martin, C.; Boucherot, K.; da Silva, C.; et al. Full-length de novo assembly of RNA-seq data in pea (Pisum sativum L.) provides a gene expression atlas and gives insights into root nodulation in this species. Plant J. 2015, 84, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Severin, A.J.; Woody, J.L.; Bolon, Y.T.; Joseph, B.; Diers, B.W.; Farmer, A.D.; Muehlbauer, G.J.; Nelson, R.X.; Grant, D.; Specht, J.E.; et al. RNA-Seq atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol. 2010, 14, 160. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.; Patel, R.K.; Tyagi, A.K.; Jain, M. De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res. 2011, 18, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Verdier, J.; Torres-Jerez, I.; Wang, M.; Andriankaja, A.; Allen, S.N.; He, J.; Tang, Y.; Murray, J.D.; Udvardi, M.K. Establishment of the Lotus japonicus Gene Expression Atlas (LjGEA) and its use to explore legume seed maturation. Plant J. 2013, 74, 351–362. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, J.A.; Iniguez, L.P.; Fu, F.; Bucciarella, B.; Miller, S.S.; Jackson, S.A.; McClean, P.E.; Li, J.; Dai, X.; Zhao, P.X.; et al. An RNA-Seq based gene expression atlas of the common bean. BMC Genom. 2014, 15, 866. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Fung-Leung, W.P.; Bittner, A.; Ngo, K.; Liu, X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS ONE 2014, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Medicago Truncatula Genome Project. Available online: http://jcvi.org/medicago/ (accessed on 23 May 2016).
- Vogel, J.P.; Garvin, D.F.; Mockler, T.C.; Schmutz, J.; Rokhsar, D.; Bevan, M.W.; Barry, K.; Lucas, S.; Harmon-Smith, M.; Lail, K. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 2010, 463, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Franssen, S.U.; Shrestha, R.P.; Bräutigam, A.; Bornberg-Bauer, E.; Weber, A.P.M. Comprehensive transcriptome analysis of the highly complex Pisum sativum genome using next generation sequencing. BMC Genom. 2011, 12, 227. [Google Scholar] [CrossRef] [PubMed]
- Duarte, J.; Rivière, N.; Baranger, A.; Aubert, G.; Burstin, J.; Cornet, L.; Lavaud, C.; Lejeune-Hénaut, I.; Martinant, J.-P.; Pichon, J.-P.; et al. Transcriptome sequencing for high throughput SNP development and genetic mapping in pea. BMC Genom. 2014, 15, 126. [Google Scholar] [CrossRef] [PubMed]
- Verma, P.; Shah, N.; Bhatia, S. Development of an expressed gene catalogue and molecular markers from the de novo assembly of short sequence reads of the lentil (Lens culinaris Medik.) transcriptome. Plant Biotechnol. J. 2013, 11, 894–905. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, S.; Bandhiwal, N.; Shah, N.; Kant, C.; Gaur, R.; Bhatia, S. Global transcriptome analysis of developing chickpea (Cicer arietinum L.) seeds. Front. Plant Sci. 2014, 5, 698. [Google Scholar] [CrossRef] [PubMed]
- Kudapa, H.; Bharti, A.K.; Cannon, S.B.; Farmer, A.D.; Mulaosmanovic, B.; Kramer, K.; Bohra, A.; Weeks, N.T.; Crow, J.A.; Tuteja, R.; et al. A Comprehensive Transcriptome Assembly of Pigeonpea (Cajanus cajan L.) using Sanger and Second-Generation Sequencing Platforms. Mol. Plant. 2012, 5, 1020–1028. [Google Scholar] [CrossRef] [PubMed]
- Ocaña, S.; Seoane, P.; Bautista, R.; Palomino, C.; Claros, G.M.; Torres, A.M.; Madrid, E. Large-Scale Transcriptome Analysis in Faba Bean (Vicia faba L.) under Ascochyta fabae Infection. PLoS ONE 2015, 10, 8e0135143. [Google Scholar] [CrossRef] [PubMed]
- Kujur, A.; Upadhyaya, H.D.; Bajaj, D.; Gowda, C.L.L.; Sharma, S.; Tyagi, A.K.; Parida, S.K. Identification of candidate genes and natural allelic variants for qtls governing plant height in chickpea. Sci. Rep. 2016, 6, 27968. [Google Scholar] [CrossRef] [PubMed]
- Saxena, M.S.; Bajaj, D.; Das, S.; Kujur, A.; Kumar, V.; Singh, M.; Bansal, K.C.; Tyagi, A.K.; Parida, S.K. An integrated genomic approach for rapid delineation of candidate genes regulating agro-morphological traits in chickpea. DNA Res. 2014. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Cogan, N.O.I.; Stephens, A.; Noy, D.; Butsch, M.; Forster, J.W.; Materne, M. EST-SNP discovery and fine-resolution genetic mapping in lentil (Lens culinaris Medik.) enables candidate gene selection for boron tolerance. Theor. Appl. Genet. 2014, 127, 703–713. [Google Scholar] [CrossRef] [PubMed]
- Fedoruk, M.J.; Vandenberg, A.; Bett, K.E. Quantitative trait loci analysis of seed quality characteristics in lentil using single nucleotide polymorphism markers. Plant Gen. 2013, 6. [Google Scholar] [CrossRef]
- Summerfield, R.J.; Roberts, E.H. Photothermal regulation of flowering in pea, lentil, faba bean and chickpea. In World Crops: Cool Season Food Legumes; Summerfield, R.J., Ed.; Springer: Berlin, Germany, 1988; pp. 911–922. [Google Scholar]
- Fratini, R.; Durán, Y.; García, P.; Pérez de la Vega, M. Identification of quantitative trait loci (QTL) for plant structure, growth habit and yield in lentil. Span J. Agric. Res. 2007, 5, 348–356. [Google Scholar] [CrossRef]
- Tullu, A.; Tar’an, B.; Warkentin, T.; Vandenberg, A. Construction of an intraspecific linkage map and QTL analysis for earliness and plant height in lentil. Crop Sci. 2008, 48, 2254–2264. [Google Scholar] [CrossRef]
- Muehlbauer, F.J.; Cho, S.; Sarkar, A.; McPhee, K.E.; Coyne, C.J.; Rajesh, P.N.; Ford, R. Application of biotechnology in breeding lentil for resistance to biotic and abiotic stress. Euphytica 2006, 147, 149–165. [Google Scholar] [CrossRef]
- PlantGDB. Available online: http://www.plantgdb.org/XGDB/phplib/download.php?GDB=Gm (accessed on 23 May 2016).
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed]
- International Chickpea Genetics and Genomics Consortium (ICGGC). Available online: http://ceg.icrisat.org/gt-bt/ICGGC/GenomeManuscript.htm (accessed on 23 May 2016).
- Varshney, R.K.; Song, C.; Saxena, R.K.; Azam, S.; Yu, S.; Sharpe, A.G.; Cannon, S.; Baek, J.; Rosen, B.D.; Taran, B.; et al. Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat. Biotechnol. 2013, 31, 240–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shendure, J.; Ji, H. Next-generation DNA sequencing. Nat Biotechnol. 2008, 26, 1135–1145. [Google Scholar] [CrossRef] [PubMed]
- Arabidopsis Genome Initiative 2000. Available online: https://www.arabidopsis.org (accessed on 23 May 2016).
- Varshney, R.K.; Chen, W.; Li, Y.; Bharti, A.K.; Saxena, R.K.; Schlueter, J.A.; Donoghue, M.T.; Azam, S.; Fan, G.; Whaley, A.M. Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat Biotechnol. 2012, 30, 83–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, H.; Mun, J.H.; Kim, D.J.; Zhu, H.; Baek, J.M.; Mudge, J.; Roe, B.; Ellis, N.; Doyle, J.; Kiss, G.B.; et al. Estimating genome conservation between crop and model legume species. Proc. Natl. Acad. Sci. USA 2004, 101, 15289–15294. [Google Scholar] [CrossRef] [PubMed]
- Shakoor, N.; Nair, R.; Crasta, O.; Morris, G.; Feltus, A.; Kresovich, S.A. Sorghum bicolor expression atlas reveals dynamic genotype-specific expression profiles for vegetative tissues of grain, sweet and bioenergy sorghums. BMC Plant Biol. 2014, 14, 35. [Google Scholar] [CrossRef] [PubMed]
- Tucker, S.C. Floral development in legumes. Plant Physiol. 2003, 131, 911–926. [Google Scholar] [CrossRef] [PubMed]
- Takano, J.; Noguchi, K.; Yasumori, M.; Kobayashi, M.; Gajdos, Z.; Miwa, K.; Hayashi, H.; Yoneyama, T.; Fujiwara, T. Arabidopsis boron transporter for xylem loading. Nature 2002, 420, 337–340. [Google Scholar] [CrossRef] [PubMed]
- Takano, J.; Wada, M.; Ludewig, U.; Schaaf, G.; von Wirén, N.; Fujiwara, T. The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell 2006, 18, 1498–1509. [Google Scholar] [CrossRef] [PubMed]
- Tyerman, S.D.; Niemietz, C.M.; Bramley, H. Plant aquaporins, multifunctional water and solute channels with expanding roles. Plant Cell Environ. 2002, 25, 173–194. [Google Scholar] [CrossRef] [PubMed]
- Bogacki, P.; Peck, D.M.; Nair, R.M.; Howie, J.; Oldach, K.H. Genetic analysis of tolerance to Boron toxicity in the legume Medicago truncatula. BMC Plant Biol. 2013, 13, 54. [Google Scholar] [CrossRef] [PubMed]
- Hecht, V.; Foucher, F.; Ferrandiz, C.; Macknight, R.; Navarro, C.; Morin, J.; Vardy, M.E.; Ellis, N.; Beltrán, J.P.; Rameau, C.; Weller, J.L. Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 2005, 137, 1420–1434. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.Y.; Shin, J.H.; Kang, Y.J.; Shim, S.R.; Lee, S.H. Divergence of flowering genes in soybean. J. Biosci. 2012, 37, 857–870. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Xie, Y.; Wu, G.; Tang, J.; Luo, R.; Patterson, J.; Liu, S.; Huang, W.; He, G.; Gu, S.; Li, S.; et al. SOAPdenovo-Trans: De novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 2014, 30, 1660–1666. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Madan, A. CAP3: A DNA sequence assembly program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef] [PubMed]
- KEGG Automatic Annotation Server (KAAS). Available online: http:/www.genome.jp/tools/kaas/ (accessed on 23 May 2016).
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997. 2013. Available online: https://arxiv.org/abs/1303.3997 (accessed on 2 November 2016).
- BatchPrimer3. Available online: http://batchprimer3.bioinformatics.ucdavis.edu/cgi-bin/batchprimer3/batchprimer3.cgi (accessed on 28 August 2016).
- Song, J.; Clemens, J.; Jameson, P.E. Quantitative expression analysis of the ABC genes in Sophora tetraptera, a woody legume with an unusual sequence of floral organ development. J. Exp. Bot. 2008, 59, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]



| Primary Assembly | Statistics |
|---|---|
| SOAPdenovo-Trans | |
| Total number of filtered reads | 660,842,789 |
| Total number of reads in contig assembly | 553,644,566 |
| Total number of scaffolds and contigs | 77,778 |
| N50 | 1731 |
| Total base pairs | 76,992,636 |
| Total base pairs without 'N' | 75,665,777 |
| Secondary assembly | |
| CAP3 | |
| Total number of scaffolds and contigs | 58,994 |
| N50 | 1719 |
| Total base pairs | 66,767,914 |
| Total base pairs without 'N' | 65,746,675 |
| Assembly | Number of Transcripts | N50 | Average Transcript Length (bp) | Number of Transcripts with BLAST Hit to Reference Assembly | Number of Unique Reference Transcripts Having Hits |
|---|---|---|---|---|---|
| Kaur et al. (2011) assembly [17] | 84,069 | 349 | 360 | 75,747 | 23,417 |
| Sharpe et al. (2013) assembly [8] | 50,146 | 530 | 501 | 48,013 | 16,905 |
| Reference assembly | 58,994 | 1719 | 1132 | - | - |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sudheesh, S.; Verma, P.; Forster, J.W.; Cogan, N.O.I.; Kaur, S. Generation and Characterisation of a Reference Transcriptome for Lentil (Lens culinaris Medik.). Int. J. Mol. Sci. 2016, 17, 1887. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17111887
Sudheesh S, Verma P, Forster JW, Cogan NOI, Kaur S. Generation and Characterisation of a Reference Transcriptome for Lentil (Lens culinaris Medik.). International Journal of Molecular Sciences. 2016; 17(11):1887. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17111887
Chicago/Turabian StyleSudheesh, Shimna, Preeti Verma, John W. Forster, Noel O. I. Cogan, and Sukhjiwan Kaur. 2016. "Generation and Characterisation of a Reference Transcriptome for Lentil (Lens culinaris Medik.)" International Journal of Molecular Sciences 17, no. 11: 1887. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17111887





