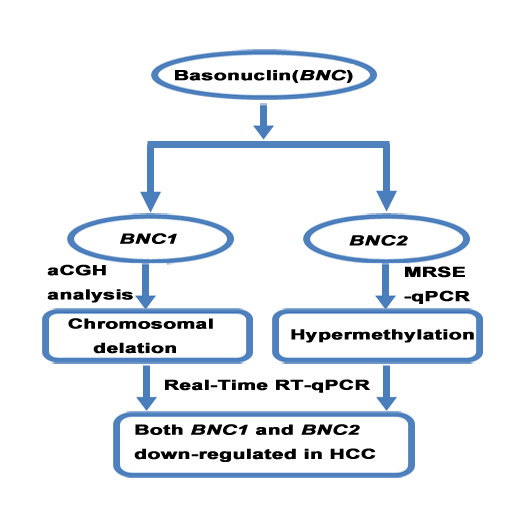
Decreased Expression of BNC1 and BNC2 Is Associated with Genetic or Epigenetic Regulation in Hepatocellular Carcinoma
Abstract
:1. Introduction
2. Results
2.1. The Expression of Basonuclin 1 (BNC1) and Basonuclin 2 (BNC2) Genes Were Down-Regulated in Hepatocellular Carcinoma (HCC) Cell Lines and Primary HCC Tissues
2.2. Chromosomal Loss of BNC1 and BNC2 Genes in Primary HCC Tumor Tissues


2.3. Promoter Methylation of BNC1 and BNC2 in HCC Cell Lines

2.4. Methylation Status of BNC1 in Primary HCC Tissues

2.5. Correlation between BNC1 Hypermethylation and Clinicopathological Features
| Feature | Hypermethylated (n = 63) | Unmethylated (n = 64) | p-Value | |
|---|---|---|---|---|
| Gender | Male | 37 | 50 | 0.019 |
| Female | 26 | 14 | ||
| Age (years) | ≥50 | 45 | 48 | 0.649 |
| <50 | 18 | 16 | ||
| AFP (μg/L) | ≥400 | 31 | 24 | 0.180 |
| <400 | 31 | 39 | ||
| Tumor diameter | ≥5cm | 48 | 47 | 0.721 |
| <5cm | 15 | 17 | ||
| Tumor number | 1 | 50 | 45 | 0.240 |
| ≥2 | 13 | 19 | ||
| Portal Vein Invasion | yes | 7 | 15 | 0.072 |
| no | 54 | 48 | ||
| BCLC stage | A | 31 | 35 | 0.536 |
| B or C | 32 | 29 | ||
3. Discussion
4. Materials and Methods
4.1. Tissue Samples and HCC Cell Lines
4.2. Gene Expression Analysis
| Clinicopathological Parameters Variables | Cases n (%) n = 127 | |
|---|---|---|
| Age | Median (Range) | 57 (11–80) |
| Gender | Male | 87 (68.50) |
| Female | 40 (31.50) | |
| Liver cirrhosis | Yes | 115 (90.55) |
| No | 10 (7.87) | |
| N/A | 2 (1.58) | |
| Portal vein tumor thrombosis | Present | 22 (17.32) |
| Absent | 102 (80.32) | |
| N/A | 3 (2.36) | |
| Tumor size | ≥5 cm | 96 (75.59) |
| <5 cm | 31 (24.41) | |
| Infection background | HBV | 94 (70.02) |
| HCV | 13 (10.24) | |
| Without HBV or HCV | 20 (15.75) | |
| Tumor encapsulation | Complete | 104 (81.89) |
| Uncomplete | 16 (12.60) | |
| BCLC stage | N/A | 7 (5.51) |
| A | 66(51.97) | |
| B | 17(13.39) | |
| C | 44(34.64) | |
| D | 0 | |
4.3. High-Density Oligonucleotide aCGH Analysis
4.4. CpG Island (CGI) Methylation Assay
4.5. Bisulfite Sequencing
4.6. 5-Aza-2′-deoxycytidine Treatment in HCC Cell Lines
4.7. Statistical Analyses
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Jiang, L.; Guan, X.Y. The genetic and epigenetic alterations in human hepatocellular carcinoma: A recent update. Protein Cell 2014, 5, 673–691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Momparler, R.L. Cancer epigenetics. Oncogene 2003, 22, 6479–6483. [Google Scholar] [CrossRef] [PubMed]
- Tseng, H.; Green, H. Basonuclin: A keratinocyte protein with multiple paired zinc fingers. Proc. Natl. Acad. Sci. USA 1992, 89, 10311–10315. [Google Scholar] [CrossRef] [PubMed]
- Romano, R.A.; Li, H.; Tummala, R.; Maul, R.; Sinha, S. Identification of Basonuclin2, a DNA-binding zinc-finger protein expressed in germ tissues and skin keratinocytes. Genomics 2004, 83, 821–833. [Google Scholar] [CrossRef] [PubMed]
- Vanhoutteghem, A.; Djian, P. Basonuclin 2: An extremely conserved homolog of the zinc finger protein basonuclin. Proc. Natl. Acad. Sci. USA 2004, 101, 3468–3473. [Google Scholar] [CrossRef] [PubMed]
- Boldrup, L.; Coates, P.J.; Laurell, G.; Nylander, K. P63 Transcriptionally regulates BNC1, a Pol I and Pol II transcription factor that regulates ribosomal biogenesis and epithelial differentiation. Eur. J. Cancer 2012, 48, 1401–1406. [Google Scholar] [CrossRef] [PubMed]
- Vanhoutteghem, A.; Djian, P. Basonuclins 1 and 2, whose genes share a common origin, are proteins with widely different properties and functions. Proc. Natl. Acad. Sci. USA 2006, 103, 12423–12428. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.M.; Guzzetta, A.A.; Bailey, V.J.; Downing, S.R.; van Neste, L.; Chiappinelli, K.B.; Keeley, B.P.; Stark, A.; Herrera, A.; Wolfgang, C.; et al. Novel methylation biomarker panel for the early detection of pancreatic cancer. Clin. Cancer Res. 2013, 19, 6544–6555. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.R.; Ricketts, C.; Gentle, D.; Abdulrahman, M.; Clarke, N.; Brown, M.; Kishida, T.; Yao, M.; Latif, F.; Maher, E.R. Identification of candidate tumour suppressor genes frequently methylated in renal cell carcinoma. Oncogene 2010, 29, 2104–2117. [Google Scholar] [CrossRef] [PubMed]
- Shames, D.S.; Girard, L.; Gao, B.; Sato, M.; Lewis, C.M.; Shivapurkar, N.; Jiang, A.; Perou, C.M.; Kim, Y.H.; Pollack, J.R.; et al. A genome-wide screen for promoter methylation in lung cancer identifies novel methylation markers for multiple malignancies. PLoS Med. 2006, 3, e486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, W.G.; Wierda, W.G.; Lin, E.; Kuang, S.Q.; Bekele, B.N.; Estrov, Z.; Wei, Y.; Yang, H.; Keating, M.J.; Garcia-Manero, G. Genome-wide DNA methylation profiling of chronic lymphocytic leukemia allows identification of epigenetically repressed molecular pathways with clinical impact. Epigenetics 2010, 5, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Pangeni, R.P.; Channathodiyil, P.; Huen, D.S.; Eagles, L.W.; Johal, B.K.; Pasha, D.; Hadjistephanou, N.; Nevell, O.; Davies, C.L.; Adewumi, A.I.; et al. The GALNT9, BNC1 and CCDC8 genes are frequently epigenetically dysregulated in breast tumours that metastasise to the brain. Clin. Epigenet. 2015, 7. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Fan, D.; Zhang, F.; Price, J.E.; Lee, J.S.; Marchetti, D.; Fidler, I.J.; Langley, R.R. Selection of brain metastasis-initiating breast cancer cells determined by growth on hard agar. Am. J. Pathol. 2011, 178, 2357–2366. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, L.C.; Hamer, M.A.; Gunn, D.A.; Deelen, J.; Lall, J.S.; van Heemst, D.; Uh, H.W.; Hofman, A.; Uitterlinden, A.G.; Griffiths, C.E.; et al. A Genome-Wide association study identifies the skin color genes IRF4, MC1R, ASIP, and BNC2 influencing facial pigmented spots. J. Investig. Dermatol. 2015, 135, 1735–1742. [Google Scholar] [CrossRef] [PubMed]
- Akagi, T.; Ito, T.; Kato, M.; Jin, Z.; Cheng, Y.; Kan, T.; Yamamoto, G.; Olaru, A.; Kawamata, N.; Boult, J.; et al. Chromosomal abnormalities and novel disease-related regions in progression from Barrett’s esophagus to esophageal adenocarcinoma. Int. J. Cancer. 2009, 125, 2349–2359. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Yang, Z.; Li, W.; Li, X.; Wang, Y.; Zhang, J.; Xu, C.; Chen, P.J.; Hou, J.; Mccrae, M.A.; et al. Re-evaluation of the carcinogenic significance of hepatitis B virus integration in hepatocarcinogenesis. PLoS ONE 2012, 7, e40363. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cheng, J.; Xu, C.; Liu, S.; Jiang, S.; Xu, Q.; Chen, X.; Zhuang, H.; Lu, F. Quantitative methylation analysis reveals gender and age differences in p16INK4a hypermethylation in hepatitis B virus-related hepatocellular carcinoma. Liver Int. 2012, 32, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Tseng, H.; Green, H. Association of basonuclin with ability of keratinocytes to multiply and with absence of terminal differentiation. J. Cell Biol. 1994, 126, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Heffelfinger, S.C.; Hawkins, H.H.; Barrish, J.; Taylor, L.; Darlington, G.J. SK HEP-1: A human cell line of endothelial origin. Vitro Cell Dev. Biol. 1992, 28A, 136–142. [Google Scholar] [CrossRef]
- Tarocchi, M.; Polvani, S.; Marroncini, G.; Galli, A. Molecular mechanism of hepatitis B virus-induced hepatocarcinogenesis. World J. Gastroenterol. 2014, 20, 11630–11640. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, J.; Cheng, J.; Ding, S.; Li, M.; Sun, S.; Zhang, L.; Liu, S.; Chen, X.; Zhuang, H.; et al. An integrated analysis of SOCS1 down-regulation in HBV infection-related hepatocellular carcinoma. J. Viral Hepat. 2014, 21, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Lu, F.M.; Hart, S.; Foroni, L.; Rabbani, H.; Hammarstrom, L.; Yuille, M.R.; Catovsky, D.; Webster, A.D.; Vorechovsky, I. Ataxia-telangiectasia and T-cell leukemias: No evidence for somatic ATM mutation in sporadic T-ALL or for hypermethylation of the ATM-NPAT/E14 bidirectional promoter in T-PLL. Cancer Res. 1998, 58, 2293–2297. [Google Scholar] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Zhang, X.; Liu, Y.; Lu, F.; Chen, X. Decreased Expression of BNC1 and BNC2 Is Associated with Genetic or Epigenetic Regulation in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2016, 17, 153. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17020153
Wu Y, Zhang X, Liu Y, Lu F, Chen X. Decreased Expression of BNC1 and BNC2 Is Associated with Genetic or Epigenetic Regulation in Hepatocellular Carcinoma. International Journal of Molecular Sciences. 2016; 17(2):153. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17020153
Chicago/Turabian StyleWu, Yali, Xiaolei Zhang, Yongzhen Liu, Fengmin Lu, and Xiangmei Chen. 2016. "Decreased Expression of BNC1 and BNC2 Is Associated with Genetic or Epigenetic Regulation in Hepatocellular Carcinoma" International Journal of Molecular Sciences 17, no. 2: 153. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17020153