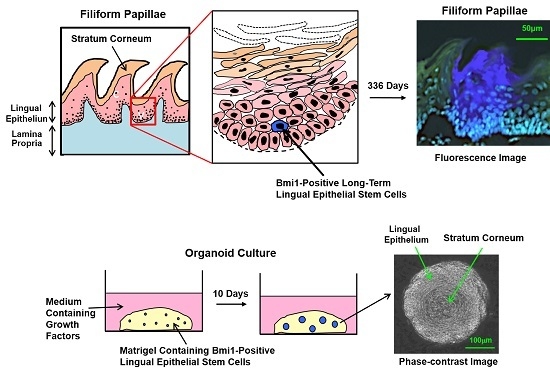
Lingual Epithelial Stem Cells and Organoid Culture of Them
Abstract
:1. Introduction
2. Lingual Stem Cell Markers
2.1. Keratin 5 and Keratin 14
| Marker | Keratin 14 | NTPDase2 | Bmi1 | Tcf3 |
|---|---|---|---|---|
| Position of Maker-Positive Cells in Lingual Epithelium | Basal Layer and Mature Keratinized Cell Layer | Basal Layer, Suprabasal Layer and Taste Bud | The 2nd or 3rd Layer from Basal Layer (1 Cell per IPP) | Basal Layer (2-5 Cells per IPP) |
| Differentiation to Papillae | Yes (Filiform and Fungiform Papillae) | Yes (Filiform, Fungiform and Circumvallate Papillae) | Yes (Filiform Papillae Only) | Yes (Filiform Papillae Only) |
| Differentiation to Taste Buds | Yes | Yes | No | (Not Described) |
| Period of Chase | 30 days | 150 days | 336 days | 180 days |
| Reference | [7] | [10] | [8] | [11] |
2.2. NTPDase2
2.3. Multicolor Lineage Tracing Method
2.4. Bmi1

2.5. Tcf3
3. Culture of Lingual Epithelial Cells
3.1. Organ Culture
3.2. Culture on Extracellular Matrix
| Culture System | Culture Setup (Day 0) Imaged | Advantage | Limitation | Ref. | |
|---|---|---|---|---|---|
| Organ Culture |  | Close to physiological conditions | Impossible to apply to adult tongues Difficult to maintain long-term growth (maximum 6 days) | [27,28,29,30] | |
| Culture on Extracellular Matrix | 2D |  | Possible to generate epithelial cell monolayer from single cells | Impossible to generate stratified keratinized epithelial cell layer | [31] |
| Organotypic Raft Culture | 3D |  | Possible to generate stratified keratinized epithelial cell layer from single cells Also possible to observe inversion activity and its process into collagen gel layer of malignant LECs | Prior preparation of the feeder layer is required Results of culture experiments are generally influenced by feeder cells | [9,32,33] |
| Organoid Culture | 3D |  | Simple and easy in culture technique Possible to generate stratified keratinized epithelial cell layer from single cells Also possible to trace the fate of each LEC | High concentration of growth factors is required | [34] |
3.3. Organotypic Raft Culture
3.4. Organoid Culture


4. Consideration of the Lingual Stem Cell Niche

5. Applications and Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hume, W.J. Kinetics of cell replacement in the stratum granulosum of mouse tongue epithelium. Cell Tissue Kinet. 1986, 19, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Hume, W.J.; Potten, C.S. The ordered columnar structure of mouse filiform papillae. J. Cell Sci. 1976, 22, 149–160. [Google Scholar] [PubMed]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, N.; Baker, N.; Stange, D.; van Es, J.H.; Abe, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Barker, N.; Huch, M.; Kujala, P.; van de Wetering, M.; Snippert, H.J.; van Es, J.H.; Sato, T.; Stange, D.E.; Begthel, H.; van den Born, M.; et al. Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 2010, 6, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Yui, S.; Nakamura, T.; Sato, T.; Nemoto, Y.; Mizutani, T.; Zheng, X.; Ichinosse, S.; Nagaishi, T.; Okamoto, R.; Tsuchiya, K.; et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nat. Med. 2012, 18, 618–623. [Google Scholar] [CrossRef] [PubMed]
- Coulombe, P.A.; Lee, C.H. Defining keratin protein function in skin epithelia: Epidermolysis bullosa simplex and its aftermath. J. Investig. Dermatol. 2012, 132, 763–775. [Google Scholar] [CrossRef] [PubMed]
- Okubo, T.; Clark, C.; Hogan, B.L.M. Cell lineage mapping of taste bud cells and keratinocytes in the mouse tongue and soft palate. Stem Cells 2009, 27, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Komai, Y.; Tokuyama, Y.; Yanai, H.; Ohe, S.; Okazaki, K.; Ueno, H. Identification of stem cells that maintain and regenerate lingual keratinized epithelial cells. Nat. Cell Biol. 2013, 15, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Okubo, T.; Randell, S.; Hogan, B.L.M. Culture of endodermal stem/progenitor cells of the mouse tongue. In Vitro Cell Dev. Biol. Anim. 2009, 45, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Cao, J.; Zhou, M. NTPDase 2+ cells generate lingual epithelia and papillae. Front. Genet. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Howard, J.M.; Nuguid, J.M.; Ngole, D.; Nguyen, H. Tcf3 expression marks both stem and progenitor cells in multiple epithelia. Development 2014, 141, 3143–3152. [Google Scholar] [CrossRef] [PubMed]
- Braun, N.; Sevigny, J.; Mishra, S.K.; Robson, S.C.; Barth, S.W.; Gerstberger, R.; Hammer, K.; Zimmermann, H. Expression of the ecto-ATPase NTPDase2 in the germinal zones of the developing and adult rat brain. Eur. J. Neurosci. 2003, 17, 1355–1364. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.L.; Sullivan, S.L.; Lavoie, E.G.; Sevigny, J.; Finger, T.E. Nucleoside triphosphate diphosphohydrolase-2 (NTPDase 2) is the ecto-ATPase of type I cells in taste buds. J. Comp. Neurol. 2006, 497, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ueno, H.; Weissman, I.L. Clonal analysis of mouse development reveals a polyclonal origin for yolk sac blood islands. Dev. Cell 2006, 11, 519–533. [Google Scholar] [CrossRef] [PubMed]
- Red-Horse, K.; Ueno, H.; Weissman, I.L.; Krasnow, M. Coronary arteries form by developmental reprogramming of venous cells. Nature 2010, 464, 549–553. [Google Scholar] [CrossRef] [PubMed]
- Rinkevich, Y.; Lindau, P.; Ueno, H.; Longaker, M.T.; Weissman, I.L. Germ-layer and lineage-restricted stem/progenitors regenerate the mouse digit tip. Nature 2011, 476, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Yanai, H.; Tanaka, T.; Ueno, H. Multicolor lineage tracing methods and intestinal tumors. J. Gastroenterol. 2013, 48, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Park, I.K.; Qian, D.; Kiel, M.; Becker, M.W.; Pihalja, M.; Weissman, I.L.; Morrison, S.J.; Clarke, M.F. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 2003, 423, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Molofsky, A.V.; Pardal, R.; Iwashita, T.; Park, I.K.; Clarke, M.F.; Morrison, S.J. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 2003, 425, 962–967. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.K.; Kim, R.H.; Kim, S.J.; Yip, F.K.; Shin, K.H.; Dimri, G.P.; Christensen, R.; Han, T.; Park, N.-H. Elevated Bmi-1 expression is associated with dysplastic cell transformation during oral carcinogenesis and is required for cancer cell replication and survival. Br. J. Cancer 2007, 96, 126–133. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Liu, Z.; Zhao, T.; Zhao, L.; Zhou, X.; Wang, A. Bmi1 drives stem-like properties and is associated with migration, invasion, and poor prognosis in tongue squamous cell carcinoma. Int. J. Biol. Sci. 2015, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sangiorgi, E.; Capecchi, M.R. Bmi1 is expressed in vivo in intestinal stem cells. Nat. Genet. 2008, 40, 915–920. [Google Scholar] [CrossRef] [PubMed]
- Komai, Y.; Tanaka, T.; Tokuyama, Y.; Yanai, H.; Ohe, S.; Omachi, T.; Atsumi, N.; Yoshida, N.; Kumano, K.; Hisha, H.; et al. Bmi1 expression in long-term germ stem cells. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Ohe, S.; Tanaka, T.; Yanai, H.; Komai, Y.; Omachi, T.; Kanno, S.; Tanaka, K.; Ishigaki, K.; Saiga, K.; Nakamura, N.; et al. Maintenance of sweat glands by stem cells located in the acral epithelium. Biochem. Biophys. Res. Commun. 2015, 466, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Yi, F.; Pereira, L.; Hoffman, J.A.; Shy, B.R.; Yuen, C.M.; Liu, D.R.; Merrill, B.J. Opposing effects of Tcf3 and Tcf1 control Wnt-stimulation of embryonic stem cell self renewal. Nat. Cell Biol. 2011, 13, 762–770. [Google Scholar] [CrossRef] [PubMed]
- Merrill, B.J.; Gat, U.; DasGupta, R.; Fuchs, E. Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes Dev. 2001, 15, 1688–1705. [Google Scholar] [CrossRef] [PubMed]
- Mbiene, J.P.; Maccallum, D.K.; Mistretta, C.M. Organ cultures of embryonic rat tongue support tongue and gustatory papilla morphogenesis in vitro without intact sensory ganglia. J. Comp. Neurol. 1997, 377, 324–340. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, H.X.; Mistretta, C.M. Bone morphogenic proteins and noggin: Inhibiting and inducing fungiform taste papilla development. Dev. Biol. 2006, 297, 198–213. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Lee, M.J.; Cho, K.W.; Lee, J.M.; Kim, Y.J.; Kim, J.Y.; Jung, H.I.; Cho, J.Y.; Cho, S.W.; Jung, H.S. Shh and ROCK1 modulate the dynamic epithelial morphogenesis in circumvallate papilla development. Dev. Biol. 2009, 325, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Sohn, W.J.; Jung, H.I.; Choi, M.A.; Han, J.H.; Gwon, G.J.; Yamamoto, H.; Lee, S.; Ryoo, Z.Y.; Park, E.K.; Shin, H.I.; et al. Reciprocal interactions of Fgf10/Fgf2b modulate the mouse tongue epithelial differentiation. Cell Tissue Res. 2011, 345, 265–273. [Google Scholar] [CrossRef]
- Ookura, T.; Kawamoto, K.; Tsuzaki, H.; Mikami, Y.; Ito, Y.; Oh, S.H.; Hino, A. Fibroblast and epidermal growth factors modulate proliferation and neural cell adhesion molecule expression in epithelial cells derived from the adult mouse tongue. In Vitro Cell Dev. Biol. Anim. 2002, 38, 365–372. [Google Scholar] [CrossRef]
- Nurmenniemi, S.; Sinikumpu, T.; Alahuhta, I.; Salo, S.; Sutinen, M.; Santala, M.; Risteli, J.; Nyberg, P.; Salo, T. A novel organotypic model mimics the tumor microenvironment. Am. J. Pathol. 2009, 175, 1281–1291. [Google Scholar] [CrossRef] [PubMed]
- Bitu, C.C.; Kauppila, J.H.; Bufalino, A.; Nurmenniemi, S.; Teppo, S.; Keinanen, M.; Vilen, S.T.; Lehenkari, P.; Nyberg, P.; Coletta, R.D.; et al. Cathepsin K is present in invasive oral tongue squamous cell carcinoma in vivo and in vitro. PLoS ONE 2013, 8, e70925. [Google Scholar]
- Hisha, H.; Tanaka, T.; Kanno, S.; Tokuyama, Y.; Komai, Y.; Ohe, S.; Yanai, H.; Omachi, T.; Ueno, H. Establishment of a novel lingual organoid culture system: Generation of organoids having mature keratinized epithelium from adult epithelial stem cells. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; van Es, J.H.; Snipper, H.J.; Strange, D.E.; Vries, R.G.; van den Born, M.; Barker, N.; Shroyer, N.F.; van de Wetering, M.; Clevers, H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 2011, 469, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Niu, C.; Ye, L.; Huang, H.; He, X.; Tong, Y.G.; Ross, J.; Haug, J.; Johnson, T.; Feng, J.O.; et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature 2003, 425, 836–841. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, T.; Kohara, H.; Noda, M.; Nagasawa, T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity 2006, 25, 977–988. [Google Scholar] [CrossRef] [PubMed]
- Kiel, M.J.; Yilmaz, O.H.; Iwashita, T.; Yilmaz, O.H.; Terhorst, C.; Morrison, S.J. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 2005, 121, 1109–1121. [Google Scholar] [CrossRef] [PubMed]
- Kinisaki, Y.; Bruns, I.; Scheiermann, C.; Ahmed, J.; Pinho, S.; Zhang, D.; Mizoguchi, T.; Wei, Q.; Lucas, D.; Ito, K.; et al. Arteriolar niches maintain haematopoietic stem cell quiescence. Nature 2013, 502, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Acar, M.; Kocherlakota, K.S.; Murphy, M.M.; Peyer, J.G.; Oguro, H.; Inra, C.N.; Jaiyeola, C.; Zhao, Z.; Luby-Phelps, K.; Morrison, S.J. Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature 2015, 526, 126–130. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, S.; Ema, H.; Karlsson, G.; Yamaguchi, T.; Miyoshi, H.; Shioda, S.; Taketo, M.M.; Karisson, S.; Iwama, A.; Nakauchi, H. Nonmyelinating Schwann cells maintain hematopoietic stem cell hibernation in the bone marrow niche. Cell 2011, 147, 1146–1158. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, L.; Pan, H.; Wang, B.; Yan, F.; Fang, X.D.; Munnee, K.; Tang, Z.G. Bmi1 essentially mediates podocalyxin-enhanced cisplatin chemoresistance in oral tongue squamous cell carcinoma. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Hayry, V.; Makinen, L.K.; Atula, T.; Sariola, H.; Makitie, A.; Leivo, I.; Keski-Santti, H.; Lundin, J.; Haglund, C.; Hagstrom, J. Bmi1 expression predicts prognosis in squamous cell carcinoma of the tongue. Br. J. Cancer 2010, 102, 892–897. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, J.; Suzuki, H.; Murata, M.; Kakei, Y.; Ri, S.; Umeda, M.; Komori, T. Clinical evaluation of application of polyglycolic acid sheet and fibrin glue spray for partial glossectomy. J. Oral Maxillofac. Surg. 2013, 71, e126–e131. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hisha, H.; Tanaka, T.; Ueno, H. Lingual Epithelial Stem Cells and Organoid Culture of Them. Int. J. Mol. Sci. 2016, 17, 168. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17020168
Hisha H, Tanaka T, Ueno H. Lingual Epithelial Stem Cells and Organoid Culture of Them. International Journal of Molecular Sciences. 2016; 17(2):168. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17020168
Chicago/Turabian StyleHisha, Hiroko, Toshihiro Tanaka, and Hiroo Ueno. 2016. "Lingual Epithelial Stem Cells and Organoid Culture of Them" International Journal of Molecular Sciences 17, no. 2: 168. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17020168