G-Protein-Coupled Lysophosphatidic Acid Receptors and Their Regulation of AKT Signaling
Abstract
:1. Introduction


2. LPA Receptors 1-3 (The EDG Family)
3. LPA Receptors 4-6 (The Non-EDG Family)
| Receptor | Species | Major Expression Sites | Biological Functions | References |
|---|---|---|---|---|
| LPA1 | Mouse | Brain, heart, lungs, stomach, kidneys, spleen, uterus, testes | Neurodevelopment regulation; neural cell proliferation, differentiation and migration; astrocyte proliferation | [1,5,20,38,55,56] |
| Human | Brain, heart, lungs, stomach, intestine, placenta, kidneys, spleen, uterus, testis | |||
| LPA2 | Mouse | Kidney, uterus, brain, testes | Cell survival; cell migration; immune system regulation | [1,5,23,43,55,56] |
| Human | Leukocytes, spleen, thymus, pancreas, brain, prostate, testes | |||
| LPA3 | Mouse | Lungs, kidney, uterus, small intestine, testes | Male and female reproductive system regulation; embryo implantation | [1,5,25,55,56] |
| Human | Heart, testes, prostate, pancreas, brain | |||
| LPA4 | Mouse | Heart, skin, ovary, thymus, lungs, kidney | Blood and lymphatic vessel development; neurite retraction; cell adhesion | [17,60,66,67,71] |
| Human | Ovary, thymus, brain, heart, testes, prostate, spleen | |||
| LPA5 | Mouse | Heart, lung, stomach, small intestine, liver, spleen, platelets, mast cells | Neurite retraction; inhibition of cell migration; calcium level regulation; water absorption; platelet activation; mast cell activation | [60,66,67,70] |
| Human | Heart, small intestine, colon, liver, spleen | |||
| LPA6 | Mouse | Hair, skin | Hair development | [66,70,77,78] |
| Human | Hair, immune cells |
4. PI3K-AKT Pathway and Its Regulation
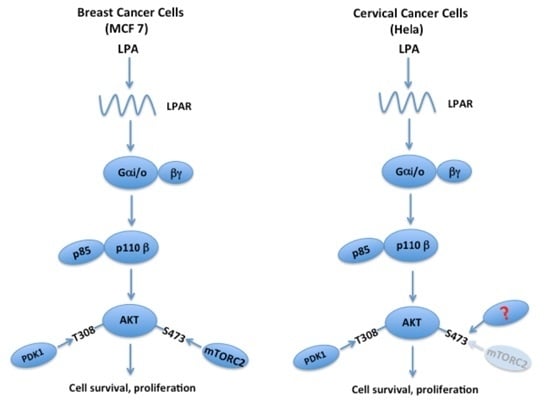
5. The LPA-PI3K-AKT Signaling Axis

6. Future Prospects
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| GPCRs | G-protein-coupled receptors |
| LPA | Lysophosphatidic acid |
| PI3K | Phosphatidyl inositol 3-kinase |
| ATX | Autotaxin |
| EDG | Endothelial differentiation genes |
| TORC2 | Target of rapamycin complex 2 |
References
- Choi, J.W.; Herr, D.R.; Noguchi, K.; Yung, Y.C.; Lee, C.W.; Mutoh, T.; Lin, M.E.; Teo, S.T.; Park, K.E.; Mosley, A.N.; et al. Lpa receptors: Subtypes and biological actions. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 157–186. [Google Scholar] [CrossRef] [PubMed]
- Moolenaar, W.H. Bioactive lysophospholipids and their G protein-coupled receptors. Exp. Cell Res. 1999, 253, 230–238. [Google Scholar] [CrossRef] [PubMed]
- Aoki, J.; Taira, A.; Takanezawa, Y.; Kishi, Y.; Hama, K.; Kishimoto, T.; Mizuno, K.; Saku, K.; Taguchi, R.; Arai, H. Serum lysophosphatidic acid is produced through diverse phospholipase pathways. J. Biol. Chem. 2002, 277, 48737–48744. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.W.; Chun, J. Lysophospholipids and their receptors in the central nervous system. Biochim. Biophys. Acta 2013, 1831, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Yung, Y.C.; Stoddard, N.C.; Chun, J. Lpa receptor signaling: Pharmacology, physiology, and pathophysiology. J. Lipid Res. 2014, 55, 1192–1214. [Google Scholar] [CrossRef] [PubMed]
- Hama, K.; Bandoh, K.; Kakehi, Y.; Aoki, J.; Arai, H. Lysophosphatidic acid (Lpa) receptors are activated differentially by biological fluids: Possible role of lpa-binding proteins in activation of lpa receptors. FEBS Lett. 2002, 523, 187–192. [Google Scholar] [CrossRef]
- Bandoh, K.; Aoki, J.; Taira, A.; Tsujimoto, M.; Arai, H.; Inoue, K. Lysophosphatidic acid (Lpa) receptors of the edg family are differentially activated by lpa species: Structure–activity relationship of cloned Lpa receptors. FEBS Lett. 2000, 478, 159–165. [Google Scholar] [CrossRef]
- Aoki, J.; Inoue, A.; Okudaira, S. Two pathways for lysophosphatidic acid production. Biochim. Biophys. Acta (BBA) 2008, 1781, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Moolenaar, W.H.; Perrakis, A. Insights into autotaxin: How to produce and present a lipid mediator. Nat. Rev. 2011, 12, 674–679. [Google Scholar] [CrossRef] [PubMed]
- Blaho, V.A.; Hla, T. Regulation of mammalian physiology, development, and disease by the sphingosine 1-phosphate and lysophosphatidic acid receptors. Chem. Rev. 2011, 111, 6299–6320. [Google Scholar] [CrossRef] [PubMed]
- Okudaira, S.; Yukiura, H.; Aoki, J. Biological roles of lysophosphatidic acid signaling through its production by autotaxin. Biochimie 2010, 92, 698–706. [Google Scholar] [CrossRef] [PubMed]
- Houben, A.J.; Moolenaar, W.H. Autotaxin and lpa receptor signaling in cancer. Cancer Metastasis Rev. 2011, 30, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Stracke, M.L.; Krutzsch, H.C.; Unsworth, E.J.; Arestad, A.; Cioce, V.; Schiffmann, E.; Liotta, L.A. Identification, purification, and partial sequence analysis of autotaxin, a novel motility-stimulating protein. J. Biol. Chem. 1992, 267, 2524–2529. [Google Scholar] [PubMed]
- Lee, H.Y.; Murata, J.; Clair, T.; Polymeropoulos, M.H.; Torres, R.; Manrow, R.E.; Liotta, L.A.; Stracke, M.L. Cloning, chromosomal localization, and tissue expression of autotaxin from human teratocarcinoma cells. Biochem. Biophys. Res. Commun. 1996, 218, 714–719. [Google Scholar] [CrossRef] [PubMed]
- Perrakis, A.; Moolenaar, W.H. Autotaxin: Structure-function and signaling. J. Lipid Res. 2014, 55, 1010–1018. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, Y.C.; Krummel, M.F.; Rosen, S.D. Autotaxin through lysophosphatidic acid stimulates polarization, motility, and transendothelial migration of naive t cells. J. Immunol. 2012, 189, 3914–3924. [Google Scholar] [CrossRef] [PubMed]
- Teo, S.T.; Yung, Y.C.; Herr, D.R.; Chun, J. Lysophosphatidic acid (lpa) in vascular development and disease. IUBMB Life 2009, 61, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, N.; Ye, X.; Chun, J. Neurobiology of lysophosphatidic acid signaling. The Neurosci.: Rev. J. Bringing Neurobiol. Neurol. Psychiatry 2002, 8, 540–550. [Google Scholar]
- Spohr, T.C.; Choi, J.W.; Gardell, S.E.; Herr, D.R.; Rehen, S.K.; Gomes, F.C.; Chun, J. Lysophosphatidic acid receptor-dependent secondary effects via astrocytes promote neuronal differentiation. J. Biol. Chem. 2008, 283, 7470–7479. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Fukushima, N.; Kingsbury, M.A.; Chun, J. Lysophosphatidic acid in neural signaling. Neuroreport 2002, 13, 2169–2175. [Google Scholar] [CrossRef] [PubMed]
- Yung, Y.C.; Mutoh, T.; Lin, M.E.; Noguchi, K.; Rivera, R.R.; Choi, J.W.; Kingsbury, M.A.; Chun, J. Lysophosphatidic acid signaling may initiate fetal hydrocephalus. Sci. Transl. Med. 2011, 3, 99ra87. [Google Scholar] [CrossRef] [PubMed]
- Yung, Y.C.; Stoddard, N.C.; Mirendil, H.; Chun, J. Lysophosphatidic acid signaling in the nervous system. Neuron 2015, 85, 669–682. [Google Scholar] [CrossRef] [PubMed]
- Knowlden, S.; Georas, S.N. The autotaxin-lpa axis emerges as a novel regulator of lymphocyte homing and inflammation. J. Immunol. 2014, 192, 851–857. [Google Scholar] [CrossRef] [PubMed]
- Gennero, I.; Laurencin-Dalicieux, S.; Conte-Auriol, F.; Briand-Mesange, F.; Laurencin, D.; Rue, J.; Beton, N.; Malet, N.; Mus, M.; Tokumura, A.; et al. Absence of the lysophosphatidic acid receptor lpa1 results in abnormal bone development and decreased bone mass. Bone 2011, 49, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Chun, J. Lysophosphatidic acid (Lpa) signaling in vertebrate reproduction. Trends Endocrinol. Metab.: TEM 2010, 21, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Sims, S.M.; Panupinthu, N.; Lapierre, D.M.; Pereverzev, A.; Dixon, S.J. Lysophosphatidic acid: A potential mediator of osteoblast-osteoclast signaling in bone. Biochim. Biophys. Acta 2013, 1831, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Tsujiuchi, T.; Araki, M.; Hirane, M.; Dong, Y.; Fukushima, N. Lysophosphatidic acid receptors in cancer pathobiology. Histol. Histopathol. 2014, 29, 313–321. [Google Scholar] [PubMed]
- Tsujiuchi, T.; Hirane, M.; Dong, Y.; Fukushima, N. Diverse effects of lpa receptors on cell motile activities of cancer cells. J. Recept. Signal Transduct. Res. 2014, 34, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Yang, W.; Zhang, Q.; Hu, Y.; Bao, L.; Damirin, A. Migration of gastric cancer cells in response to lysophosphatidic acid is mediated by lpa receptor 2. Oncol. Lett. 2013, 5, 1048–1052. [Google Scholar] [PubMed]
- Sokolov, E.; Eheim, A.L.; Ahrens, W.A.; Walling, T.L.; Swet, J.H.; McMillan, M.T.; Simo, K.A.; Thompson, K.J.; Sindram, D.; McKillop, I.H. Lysophosphatidic acid receptor expression and function in human hepatocellular carcinoma. J. Surg. Res. 2013, 180, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Popnikolov, N.K.; Dalwadi, B.H.; Thomas, J.D.; Johannes, G.J.; Imagawa, W.T. Association of autotaxin and lysophosphatidic acid receptor 3 with aggressiveness of human breast carcinoma. Tumour Biol.: J. Int. Soc. Oncodev. Biol. Med. 2012, 33, 2237–2243. [Google Scholar] [CrossRef] [PubMed]
- Komachi, M.; Sato, K.; Tobo, M.; Mogi, C.; Yamada, T.; Ohta, H.; Tomura, H.; Kimura, T.; Im, D.S.; Yanagida, K.; et al. Orally active lysophosphatidic acid receptor antagonist attenuates pancreatic cancer invasion and metastasis in vivo. Cancer Sci. 2012, 103, 1099–1104. [Google Scholar] [CrossRef] [PubMed]
- Sako, A.; Kitayama, J.; Shida, D.; Suzuki, R.; Sakai, T.; Ohta, H.; Nagawa, H. Lysophosphatidic acid (Lpa)-induced vascular endothelial growth factor (Vegf) by mesothelial cells and quantification of host-derived vegf in malignant ascites. J. Surg. Res. 2006, 130, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.I.; Chen, C.N.; Huang, M.T.; Lee, S.J.; Lin, C.H.; Chang, C.C.; Lee, H. Lysophosphatidic acid up-regulates vascular endothelial growth factor-c and lymphatic marker expressions in human endothelial cells. Cell. Mol. Life Sci. 2008, 65, 2740–2751. [Google Scholar] [CrossRef] [PubMed]
- Samadi, N.; Bekele, R.; Capatos, D.; Venkatraman, G.; Sariahmetoglu, M.; Brindley, D.N. Regulation of lysophosphatidate signaling by autotaxin and lipid phosphate phosphatases with respect to tumor progression, angiogenesis, metastasis and chemo-resistance. Biochimie 2011, 93, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Sui, Y.; Yang, Y.; Wang, J.; Li, Y.; Ma, H.; Cai, H.; Liu, X.; Zhang, Y.; Wang, S.; Li, Z.; et al. Lysophosphatidic acid inhibits apoptosis induced by cisplatin in cervical cancer cells. BioMed Res. Int. 2015, 2015, 598386. [Google Scholar] [CrossRef] [PubMed]
- Venkatraman, G.; Benesch, M.G.; Tang, X.; Dewald, J.; McMullen, T.P.; Brindley, D.N. Lysophosphatidate signaling stabilizes nrf2 and increases the expression of genes involved in drug resistance and oxidative stress responses: Implications for cancer treatment. FASEB J. Off. Publ. Federation Am. Soc. Exp. Biol. 2015, 29, 772–785. [Google Scholar] [CrossRef] [PubMed]
- Sheng, X.; Yung, Y.C.; Chen, A.; Chun, J. Lysophosphatidic acid signalling in development. Development 2015, 142, 1390–1395. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, N.; Ishii, S.; Tsujiuchi, T.; Kagawa, N.; Katoh, K. Comparative analyses of lysophosphatidic acid receptor-mediated signaling. Cell. Mol. Life Sci. CMLS 2015, 72, 2377–2394. [Google Scholar] [CrossRef] [PubMed]
- Xiang, S.Y.; Dusaban, S.S.; Brown, J.H. Lysophospholipid receptor activation of rhoa and lipid signaling pathways. Biochim. Biophys. Acta 2013, 1831, 213–222. [Google Scholar] [CrossRef] [PubMed]
- van Leeuwen, F.N.; Olivo, C.; Grivell, S.; Giepmans, B.N.G.; Collard, J.G.; Moolenaar, W.H. Rac activation by lysophosphatidic acid lpa1receptors through the guanine nucleotide exchange factor tiam1. J. Biol. Chem. 2003, 278, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Yu, S.; LaPushin, R.; Lu, Y.; Furui, T.; Penn, L.Z.; Stokoe, D.; Erickson, J.R.; Bast, R.C., Jr.; Mills, G.B. Lysophosphatidic acid prevents apoptosis in fibroblasts via g(I)-protein-mediated activation of mitogen-activated protein kinase. Biochem. J. 2000, 352 Pt 1, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Ishii, I.; Kingsbury, M.A.; Chun, J. Lysophosphatidic acid as a novel cell survival/apoptotic factor. Biochim. Biophys. Acta 2002, 1585, 108–113. [Google Scholar] [CrossRef]
- Guillermet-Guibert, J.; Bjorklof, K.; Salpekar, A.; Gonella, C.; Ramadani, F.; Bilancio, A.; Meek, S.; Smith, A.J.; Okkenhaug, K.; Vanhaesebroeck, B. The p110beta isoform of phosphoinositide 3-kinase signals downstream of g protein-coupled receptors and is functionally redundant with p110gamma. Proc. Natl. Acad. Sci. USA 2008, 105, 8292–8297. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, J.P. Lysophosphatidic acid induces inositol phosphate and calcium signals in exocrine cells from the avian nasal salt gland. J. Membarin Biol. 1995, 144, 49–58. [Google Scholar] [CrossRef]
- Gennero, I.; Xuereb, J.-M.; Simon, M.-F.; Girolami, J.-P.; Bascands, J.-L.; Chap, H.; Boneu, B.; Sié, P. Effects of lysophosphatidic acid on proliferation and cytosolic ca++ of human adult vascular smooth muscle cells in culture. Thromb. Res. 1999, 94, 317–326. [Google Scholar] [CrossRef]
- Litosch, I. Phosphatidic acid potentiates gαq stimulation of phospholipase c-β1 signaling. Biochem. Biophys. Res. Commun. 2009, 390, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-W.; Rivera, R.; Dubin, A.E.; Chun, J. Lpa4/gpr23 is a lysophosphatidic acid (Lpa) receptor utilizing gs-, gq/gi-mediated calcium signaling and g12/13-mediated rho activation. J. Biol. Chem. 2007, 282, 4310–4317. [Google Scholar] [CrossRef] [PubMed]
- Wittpoth, C.; Scholich, K.; Yigzaw, Y.; Stringfield, T.M.; Patel, T.B. Regions on adenylyl cyclase that are necessary for inhibition of activity by βγ and g(Iα) subunits of heterotrimeric g proteins. Proc. Natl. Acad. Sci. USA 1999, 96, 9551–9556. [Google Scholar] [CrossRef] [PubMed]
- Takuwa, Y.; Takuwa, N.; Sugimoto, N. The edg family g protein-coupled receptors for lysophospholipids: Their signaling properties and biological activities. J. Biochem. 2002, 131, 767–771. [Google Scholar] [CrossRef] [PubMed]
- Hecht, J.H.; Weiner, J.A.; Post, S.R.; Chun, J. Ventricular zone gene-1 (vzg-1) encodes a lysophosphatidic acid receptor expressed in neurogenic regions of the developing cerebral cortex. J. Cell Biol. 1996, 135, 1071–1083. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-J.; Van Brocklyn, J.R.; Thangada, S.; Liu, C.H.; Hand, A.R.; Menzeleev, R.; Spiegel, S.; Hla, T. Sphingosine-1-phosphate as a ligand for the g protein-coupled receptor edg-1. Science 1998, 279, 1552–1555. [Google Scholar] [CrossRef] [PubMed]
- Contos, J.J.A.; Chun, J. Genomic characterization of the lysophosphatidic acid receptor gene, lpa2/edg4, and identification of a frameshift mutation in a previously characterized cdna. Genomics 2000, 64, 155–169. [Google Scholar] [CrossRef] [PubMed]
- Bandoh, K.; Aoki, J.; Hosono, H.; Kobayashi, S.; Kobayashi, T.; Murakami-Murofushi, K.; Tsujimoto, M.; Arai, H.; Inoue, K. Molecular cloning and characterization of a novel human g-protein-coupled receptor, edg7, for lysophosphatidic acid. J. Biol. Chem. 1999, 274, 27776–27785. [Google Scholar] [CrossRef] [PubMed]
- Archbold, J.K.; Martin, J.L.; Sweet, M.J. Towards selective lysophospholipid gpcr modulators. Trends Pharmacol. Sci. 2014, 35, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.W.; Lee, C.W.; Chun, J. Biological roles of lysophospholipid receptors revealed by genetic null mice: An update. Biochim. Biophys. Acta 2008, 1781, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Anliker, B.; Choi, J.W.; Lin, M.E.; Gardell, S.E.; Rivera, R.R.; Kennedy, G.; Chun, J. Lysophosphatidic acid (lpa) and its receptor, lpa1, influence embryonic schwann cell migration, myelination, and cell-to-axon segregation. Glia 2013, 61, 2009–2022. [Google Scholar] [CrossRef] [PubMed]
- Sakai, N.; Chun, J.; Duffield, J.S.; Wada, T.; Luster, A.D.; Tager, A.M. Lpa1-induced cytoskeleton reorganization drives fibrosis through ctgf-dependent fibroblast proliferation. FASEB J. Off. Publ. Federation Am. Soc. Exp. Biol. 2013, 27, 1830–1846. [Google Scholar] [CrossRef]
- Chrencik, J.E.; Roth, C.B.; Terakado, M.; Kurata, H.; Omi, R.; Kihara, Y.; Warshaviak, D.; Nakade, S.; Asmar-Rovira, G.; Mileni, M.; et al. Crystal structure of antagonist bound human lysophosphatidic acid receptor 1. Cell 2015, 161, 1633–1643. [Google Scholar] [CrossRef] [PubMed]
- Ohuchi, H.; Hamada, A.; Matsuda, H.; Takagi, A.; Tanaka, M.; Aoki, J.; Arai, H.; Noji, S. Expression patterns of the lysophospholipid receptor genes during mouse early development. Dev. Dyn. 2008, 237, 3280–3294. [Google Scholar] [CrossRef] [PubMed]
- Contos, J.J.; Ishii, I.; Fukushima, N.; Kingsbury, M.A.; Ye, X.; Kawamura, S.; Brown, J.H.; Chun, J. Characterization of Lpa(2) (Edg4) and Lpa(1)/Lpa(2) (Edg2/Edg4) lysophosphatidic acid receptor knockout mice: Signaling deficits without obvious phenotypic abnormality attributable to Lpa(2). Mol. Cell. Biol. 2002, 22, 6921–6929. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Lai, Y.J.; Lin, W.C.; Lin, F.T. Trip6 enhances lysophosphatidic acid-induced cell migration by interacting with the lysophosphatidic acid 2 receptor. J. Biol. Chem. 2004, 279, 10459–10468. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.J.; Chen, C.S.; Lin, W.C.; Lin, F.T. C-src-mediated phosphorylation of trip6 regulates its function in lysophosphatidic acid-induced cell migration. Mol. Cell. Biol. 2005, 25, 5859–5868. [Google Scholar] [CrossRef] [PubMed]
- Komachi, M.; Tomura, H.; Malchinkhuu, E.; Tobo, M.; Mogi, C.; Yamada, T.; Kimura, T.; Kuwabara, A.; Ohta, H.; Im, D.S.; et al. Lpa1 receptors mediate stimulation, whereas lpa2 receptors mediate inhibition, of migration of pancreatic cancer cells in response to lysophosphatidic acid and malignant ascites. Carcinogenesis 2009, 30, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Hama, K.; Aoki, J. Lpa3, a unique g protein-coupled receptor for lysophosphatidic acid. Prog. Lipid Res. 2010, 49, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Yanagida, K.; Ishii, S. Non-edg family lpa receptors: The cutting edge of lpa research. J. Biochem. 2011, 150, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Ishii, S.; Noguchi, K.; Yanagida, K. Non-edg family lysophosphatidic acid (lpa) receptors. Prostaglandins Lipid Mediat. 2009, 89, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Pamuklar, Z.; Lee, J.S.; Cheng, H.-Y.; Panchatcharam, M.; Steinhubl, S.; Morris, A.J.; Charnigo, R.; Smyth, S.S. Individual heterogeneity in platelet response to lysophosphatidic acid: Evidence for a novel inhibitory pathway. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, K.; Ishii, S.; Shimizu, T. Identification of p2y9/gpr23 as a novel g protein-coupled receptor for lysophosphatidic acid, structurally distant from the edg family. J. Biol. Chem. 2003, 278, 25600–25606. [Google Scholar] [CrossRef] [PubMed]
- Yanagida, K.; Kurikawa, Y.; Shimizu, T.; Ishii, S. Current progress in non-edg family lpa receptor research. Biochim. Biophys. Acta 2013, 1831, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Sumida, H.; Noguchi, K.; Kihara, Y.; Abe, M.; Yanagida, K.; Hamano, F.; Sato, S.; Tamaki, K.; Morishita, Y.; Kano, M.R.; et al. Lpa4 regulates blood and lymphatic vessel formation during mouse embryogenesis. Blood 2010, 116, 5060–5070. [Google Scholar] [CrossRef] [PubMed]
- Yanagida, K.; Ishii, S.; Hamano, F.; Noguchi, K.; Shimizu, T. Lpa4/p2y9/gpr23 mediates rho-dependent morphological changes in a rat neuronal cell line. J. Biol. Chem. 2007, 282, 5814–5824. [Google Scholar] [CrossRef] [PubMed]
- Lee, Z.; Cheng, C.-T.; Zhang, H.; Subler, M.A.; Wu, J.; Mukherjee, A.; Windle, J.J.; Chen, C.-K.; Fang, X. Role of lpa(4)/p2y9/gpr23 in negative regulation of cell motility. Mol. Biol. Cell 2008, 19, 5435–5445. [Google Scholar] [CrossRef] [PubMed]
- Yanagida, K.; Masago, K.; Nakanishi, H.; Kihara, Y.; Hamano, F.; Tajima, Y.; Taguchi, R.; Shimizu, T.; Ishii, S. Identification and characterization of a novel lysophosphatidic acid receptor, p2y5/lpa6. J. Biol. Chem. 2009, 284, 17731–17741. [Google Scholar] [CrossRef] [PubMed]
- Pasternack, S.M.; von Kugelgen, I.; Al Aboud, K.; Lee, Y.A.; Ruschendorf, F.; Voss, K.; Hillmer, A.M.; Molderings, G.J.; Franz, T.; Ramirez, A.; et al. G protein-coupled receptor p2y5 and its ligand lpa are involved in maintenance of human hair growth. Nat. Genet. 2008, 40, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Araki, M.; Kitayoshi, M.; Dong, Y.; Hirane, M.; Ozaki, S.; Mori, S.; Fukushima, N.; Honoki, K.; Tsujiuchi, T. Inhibitory effects of lysophosphatidic acid receptor-5 on cellular functions of sarcoma cells. Growth Fact. 2014, 32, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Hirane, M.; Araki, M.; Fukushima, N.; Tsujiuchi, T. Lysophosphatidic acid receptor-5 negatively regulates cellular responses in mouse fibroblast 3t3 cells. Biochem. Biophys. Res. Commun. 2014, 446, 585–589. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Fujiwara, Y.; Liu, J.; Yue, J.; Shimizu, Y.; Norman, D.D.; Wang, Y.; Tsukahara, R.; Szabo, E.; Patil, R.; et al. Autotaxin and lpa1 and lpa5 receptors exert disparate functions in tumor cells versus the host tissue microenvironment in melanoma invasion and metastasis. Mol. Cancer Res. MCR 2015, 13, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Choi, S.; Hallden, G.; Yo, S.J.; Schichnes, D.; Aponte, G.W. P2y5 is a g(alpha)i, g(alpha)12/13 g protein-coupled receptor activated by lysophosphatidic acid that reduces intestinal cell adhesion. Am. J. Physiol. Gastrointest. Liver Physiol. 2009, 297, G641–G654. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cbioportal. Sci. Signal. 2013, 6, pl1–pl1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schultze, S.M.; Jensen, J.; Hemmings, B.A.; Tschopp, O.; Niessen, M. Promiscuous affairs of pkb/akt isoforms in metabolism. Arch. Physiol. Biochem. 2011, 117, 70–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, H.; Littlewood, T.; Bennett, M. Akt isoforms in vascular disease. Vasc. Pharmacol. 2015, 71, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Dibble, C.C.; Manning, B.D. A molecular link between akt regulation and chemotherapeutic response. Cancer Cell 2009, 16, 178–180. [Google Scholar] [CrossRef] [PubMed]
- Newton, A.C.; Trotman, L.C. Turning off akt: Phlpp as a drug target. Annu. Rev. Pharmacol. Toxicol. 2014, 54, 537–558. [Google Scholar] [CrossRef] [PubMed]
- Vanhaesebroeck, B.; Guillermet-Guibert, J.; Graupera, M.; Bilanges, B. The emerging mechanisms of isoform-specific pi3k signalling. Nat. Rev. Mol. Cell Biol. 2010, 11, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Sarbassov, D.D.; Guertin, D.A.; Ali, S.M.; Sabatini, D.M. Phosphorylation and regulation of akt/pkb by the rictor-mtor complex. Science 2005, 307, 1098–1101. [Google Scholar] [CrossRef] [PubMed]
- Jacinto, E.; Facchinetti, V.; Liu, D.; Soto, N.; Wei, S.; Jung, S.Y.; Huang, Q.; Qin, J.; Su, B. Sin1/mip1 maintains rictor-mtor complex integrity and regulates akt phosphorylation and substrate specificity. Cell 2006, 127, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Song, M.S.; Salmena, L.; Pandolfi, P.P. The functions and regulation of the pten tumour suppressor. Nat. Rev. Mol. Cell Biol. 2012, 13, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Zeller, K.S.; Idevall-Hagren, O.; Stefansson, A.; Velling, T.; Jackson, S.P.; Downward, J.; Tengholm, A.; Johansson, S. Pi3-kinase p110α mediates β1 integrin-induced akt activation and membrane protrusion during cell attachment and initial spreading. Cell Signal. 2010, 22, 1838–1848. [Google Scholar] [CrossRef] [PubMed]
- Riaz, A.; Zeller, K.S.; Johansson, S. Receptor-specific mechanisms regulate phosphorylation of akt at ser473: Role of rictor in β1 integrin-mediated cell survival. PLoS ONE 2012, 7, e32081. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Viciana, P.; Sabatier, C.; McCormick, F. Signaling specificity by ras family gtpases is determined by the full spectrum of effectors they regulate. Mol. Cell. Biol. 2004, 24, 4943–4954. [Google Scholar] [CrossRef] [PubMed]
- Fritsch, R.; de Krijger, I.; Fritsch, K.; George, R.; Reason, B.; Kumar, M.S.; Diefenbacher, M.; Stamp, G.; Downward, J. Ras and rho families of gtpases directly regulate distinct phosphoinositide 3-kinase isoforms. Cell 2013, 153, 1050–1063. [Google Scholar] [CrossRef] [PubMed]
- Santi, S.A.; Lee, H. The akt isoforms are present at distinct subcellular locations. Am. J. Physiol. 2010, 298, C580–C591. [Google Scholar] [CrossRef] [PubMed]
- Villagrasa, P.; Diaz, V.M.; Vinas-Castells, R.; Peiro, S.; Del Valle-Perez, B.; Dave, N.; Rodriguez-Asiain, A.; Casal, J.I.; Lizcano, J.M.; Dunach, M.; et al. Akt2 interacts with snail1 in the e-cadherin promoter. Oncogene 2012, 31, 4022–4033. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, M.; Onishi, K.; Kikuchi, C.; Gotoh, Y. Scaffolding function of pak in the pdk1-akt pathway. Nat. Cell Biol. 2008, 10, 1356–1364. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Naito, M.; Tsuruo, T.; Fujita, N. Freud-1/aki1, a novel pdk1-interacting protein, functions as a scaffold to activate the pdk1/akt pathway in epidermal growth factor signaling. Mol. Cell. Biol. 2008, 28, 5996–6009. [Google Scholar] [CrossRef] [PubMed]
- Luan, B.; Zhao, J.; Wu, H.; Duan, B.; Shu, G.; Wang, X.; Li, D.; Jia, W.; Kang, J.; Pei, G. Deficiency of a β-arrestin-2 signal complex contributes to insulin resistance. Nature 2009, 457, 1146–1149. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.-H.; Jo, U.; Kohrman, A.; Rezaeian, A.H.; Chou, P.-C.; Logothetis, C.; Lin, H.-K. Posttranslational regulation of akt in human cancer. Cell Biosci. 2014, 4, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.H.; Jono, H.; Komatsu, K.; Woo, C.-H.; Lee, J.; Miyata, M.; Matsuno, T.; Xu, X.; Huang, Y.; Zhang, W.; et al. Cyld negatively regulates transforming growth factor-β-signalling via deubiquitinating akt. Nat. Commun. 2012, 3, 771. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.C.; Kim, K.M.; Lee, K.S.; Namkoong, S.; Lee, S.J.; Han, J.A.; Jeoung, D.; Ha, K.S.; Kwon, Y.G.; Kim, Y.M. Serum bioactive lysophospholipids prevent trail-induced apoptosis via pi3k/akt-dependent cflip expression and bad phosphorylation. Cell Death Differ. 2004, 11, 1287–1298. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.K.; Yun, S.J.; Do, K.H.; Kim, M.S.; Cho, M.; Suh, D.S.; Kim, C.D.; Kim, J.H.; Birnbaum, M.J.; Bae, S.S. Lysophosphatidic acid induces cell migration through the selective activation of akt1. Exp. Mol. Med. 2008, 40, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Gonzalez, M.I.; Meinkoth, J.L.; Field, J.; Kazanietz, M.G.; Tennekoon, G.I. Lysophosphatidic acid promotes survival and differentiation of rat schwann cells. J.Biol. Chem. 2003, 278, 9585–9591. [Google Scholar] [CrossRef] [PubMed]
- Murga, C.; Fukuhara, S.; Gutkind, J.S. A novel role for phosphatidylinositol 3-kinase beta in signaling from g protein-coupled receptors to akt. J. Biol. Chem. 2000, 275, 12069–12073. [Google Scholar] [CrossRef] [PubMed]
- Baudhuin, L.M.; Cristina, K.L.; Lu, J.; Xu, Y. Akt activation induced by lysophosphatidic acid and sphingosine-1-phosphate requires both mitogen-activated protein kinase kinase and p38 mitogen-activated protein kinase and is cell-line specific. Mol. Pharmacol. 2002, 62, 660–671. [Google Scholar] [CrossRef] [PubMed]
- Kihara, Y.; Mizuno, H.; Chun, J. Lysophospholipid receptors in drug discovery. Exp. Cell Res. 2015, 333, 171–177. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riaz, A.; Huang, Y.; Johansson, S. G-Protein-Coupled Lysophosphatidic Acid Receptors and Their Regulation of AKT Signaling. Int. J. Mol. Sci. 2016, 17, 215. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17020215
Riaz A, Huang Y, Johansson S. G-Protein-Coupled Lysophosphatidic Acid Receptors and Their Regulation of AKT Signaling. International Journal of Molecular Sciences. 2016; 17(2):215. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17020215
Chicago/Turabian StyleRiaz, Anjum, Ying Huang, and Staffan Johansson. 2016. "G-Protein-Coupled Lysophosphatidic Acid Receptors and Their Regulation of AKT Signaling" International Journal of Molecular Sciences 17, no. 2: 215. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17020215