Transcriptome-Based Identification of Differently Expressed Genes from Xanthomonas oryzae pv. oryzae Strains Exhibiting Different Virulence in Rice Varieties
Abstract
:1. Introduction
2. Results
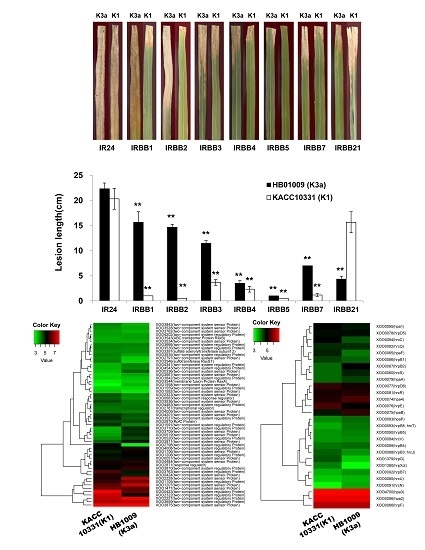
2.1. Virulence of the Xoo K1 Race and K3a Race Strains
2.2. RNA-seq Reads Mapping and GO Term Enrichment of Differentially Expressed Genes
2.3. Differential Expression of hrp Genes between the Xoo K1 Race and K3a Race Strains
2.4. Differential Expression of Two-Component Systems between the Xoo K1 Race and K3a Race Strains
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Virulence Assay
4.3. Total RNA Isolation, Illumina Sequencing, and Data Analysis
4.4. Quantitative Real-Time RT-PCR Assay
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| Xoo | Xanthomonasoryzae pv. oryzae |
| BB | bacterial blight |
| hrp | hypersensitive response and pathogenicity |
| TCSs | two-component regulatory systems |
| PRRs | pattern recognition receptors |
| QS | quorum sensing |
| DSF | diffusible signal factor |
| rax | required for activation of XA21-mediated immunity |
| TTSS | type III secretion system |
| DEGs | differentially expressed genes |
| qRT-PCR | quantitative real-time RT-PCR |
References
- Mew, T.W. Current status and future prospects of research on bacterial blight of rice. Annu. Rev. Phytopathol. 1987, 25, 359–382. [Google Scholar] [CrossRef]
- Bhasin, H.; Bhatia, D.; Raghuvanshi, S.; Lore, J.S.; Sahi, G.K.; Kaur, B.; Vikal, Y.; Singh, K. New PCR-based sequence-tagged site marker for bacterial blight resistance gene Xa38 of rice. Mol. Breed. 2012, 30, 607–611. [Google Scholar] [CrossRef]
- Natrajkumar, P.; Sujatha, K.; Laha, G.S.; Srinivasarao, K.; Mishra, B.; Viraktamath, B.C.; Hari, Y.; Reddy, C.S.; Balachandran, S.M.; Ram, T.; et al. Identification and fine-mapping of Xa33, a novel gene for resistance to Xanthomonas oryzae pv. oryzae. Phytopathology 2012, 102, 222–228. [Google Scholar]
- Song, W.Y.; Pi, L.Y.; Wang, G.L.; Gardner, J.; Holstion, T.; Ronald, P.C. Evolution of the rice Xa21 disease resistance gene family. Plant Cell 1997, 9, 1279–1287. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Sanchez, A.; Khush, G.S.; Zhu, Y.; Huang, N. Construction of a BAC contig containing the xa5 locus in rice. Theor. Appl. Genet. 1998, 97, 1120–1124. [Google Scholar] [CrossRef]
- Sun, X.; Yang, Z.; Wang, S.; Zhang, Q. Identification of a 47 kb DNA fragment containing Xa4, a locus for bacterial blight resistance in rice. Theor. Appl. Genet. 2003, 106, 683–687. [Google Scholar] [PubMed]
- Gu, K.; Yang, B.; Tian, D.; Wu, L.; Wang, D.; Sreekala, C.; Yang, F.; Chu, Z.; Wang, G.L.; White, F.F.; et al. R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 2005, 435, 1122–1125. [Google Scholar] [CrossRef] [PubMed]
- Niño-Liu, D.O.; Ronald, P.C.; Bogdanove, A.J. Xanthomonas oryzae pathovars: Model pathogens of a model crop. Mol. Plant Pathol. 2006, 7, 303–324. [Google Scholar] [CrossRef] [PubMed]
- Cheema, K.; Grewal, N.; Vikal, Y.; Sharma, R.; Lore, J.S.; Das, A.; Bhatia, D.; Mahajan, R.; Gupta, V.; Bharaj, T.S.; et al. A novel bacterial blight resistance gene from Oryza nivara mapped to 38 kb region on chromosome 4L and transferred to Oryza sativa L. Genet. Res. 2008, 90, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Tian, D.; Wang, J.; Zeng, X.; Gu, K.; Qiu, C.; Yang, X.; Zhou, Z.; Goh, M.; Luo, Y.; Murata-Hori, M.; et al. The rice TAL effector–dependent resistance protein XA10 triggers cell death and calcium depletion in the endoplasmic reticulum. Plant Cell 2014, 26, 497–515. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, X.; Fan, Y.; Gao, Y.; Zhu, Q.; Zheng, C.; Qin, T.; Li, Y.; Che, J.; Zhang, M.; et al. XA23 is an executor R protein and confers broad-spectrum disease resistance in rice. Mol. Plant 2015, 8, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Khush, G.S.; Bacalangco, E.; Ogawa, T. A new gene for resistance to bacterial blight from O. longistaminata. Rice Genet. Newsl. 1990, 7, 121–122. [Google Scholar]
- Shen, Y.; Sharma, P.; da Silva, F.G.; Ronald, P. The Xanthomonas oryzae pv. oryzae raxP and raxQ genes encode an ATP sulfurylase and adenosine-5’-phosphosulphate kinase that are required for AvrXa21 avirulence activity. Mol. Microbiol. 2002, 44, 37–48. [Google Scholar] [PubMed]
- Burdman, S.; Shen, Y.; Lee, S.W.; Xue, Q.; Ronald, P.C. RaxH/RaxR: A two-component regulatory system in Xanthomonas oryzae pv. oryzae required for AvrXa21 activity. Mol. Plant Microbe Interact. 2004, 17, 602–612. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, F.G.; Shen, Y.; Dardick, C.; Burdman, S.; Yadav, R.C.; de Leon, A.L.; Ronald, P.C. Bacterial genes involved in type I secretion and sulfation are required to elicit the rice Xa21-mediated innate immune response. Mol. Plant Microbe Interact. 2004, 17, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.; Han, S.W.; Sririyanum, M.; Park, C.J.; Seo, Y.S.; Ronald, P.C. A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity. Science 2009, 326, 850–853. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.; Han, S.W.; Sririyanum, M.; Park, C.J.; Seo, Y.S.; Ronald, P.C. Retraction. A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity. Science 2013, 342, 191. [Google Scholar] [CrossRef] [PubMed]
- Yun, M.S.; Lee, E.J.; Cho, Y.S. Pathogenic specialization of the rice bacterial leaf blight pathogen, Xanthomonas campestris pv. oryzae: Race classification based on reactions of Korean differential varieties. Korean J. Appl. Entomol. 1985, 24, 97–101. [Google Scholar]
- Shin, M.S.; Shin, H.T.; Jun, B.T.; Choi, B.S. Effect of inoculation of compatible and incompatible bacterial blight races on grain yield and quality of two rice cultivars. Korean J. Breed. Sci. 1992, 24, 264–267. [Google Scholar]
- Noh, T.H.; Lee, D.K.; Park, J.C.; Shim, H.K.; Choi, M.Y.; Kang, M.H.; Kim, J.D. Effect of bacterial leaf blight occurrence on rice yield and grain quality in different rice growth stage. Res. Plant Dis. 2007, 13, 20–23. [Google Scholar] [CrossRef]
- Kim, K.Y.; Shin, M.S.; Kim, W.J.; Mo, Y.J.; Nam, J.K.; Noh, T.H.; Kim, B.K.; Ko, J.K. Effective combination of resistance genes against rice bacterial blight pathogen. Korean J. Breed. Sci. 2009, 41, 244–251. [Google Scholar]
- Noh, T.H.; Lee, D.K.; Kang, M.H.; Shin, M.S.; Na, S.Y. Identification of new race of Xanthomonas oryzae pv. oryzae (Xoo) in Korea. Phytopathology 2003, 93, S66. [Google Scholar]
- Song, E.S.; Kim, S.Y.; Noh, T.H.; Cho, H.; Chae, S.C.; Lee, B.M. PCR-based assay for rapid and specific detection of the new Xanthomonas oryzae pv. oryzae K3a race using an AFLP-derived marker. J. Microbiol. Biotechnol. 2014, 24, 732–739. [Google Scholar] [PubMed]
- Lee, B.M.; Park, Y.J.; Park, D.S.; Kang, H.W.; Kim, J.G.; Song, E.S.; Park, I.C.; Yoon, U.H.; Hahn, J.H.; Koo, B.S.; et al. The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 2005, 33, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.G.; Park, B.K.; Yoo, C.H.; Jeon, E.; Oh, J.; Hwang, I. Characterization of the Xanthomonas axonopodis pv. glycines Hrp pathogenicity island. J. Bacteriol. 2003, 185, 3155–3166. [Google Scholar] [PubMed]
- Cho, H.J.; Park, Y.J.; Noh, T.H.; Kim, Y.T.; Kim, J.G.; Song, E.S.; Lee, D.H.; Lee, B.M. Molecular analysis of the hrp gene cluster in Xanthomonas oryzae pathovar oryzae KACC10859. Microb. Pathog. 2008, 44, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Wengelnik, K.; Rossier, O.; Bonas, U. Mutations in the regulatory gene hrpG of Xanthomonas campestris pv. vesicatoria result in constitutive expression of all hrp genes. J. Bacteriol. 1999, 181, 6828–6831. [Google Scholar] [PubMed]
- Wengelnik, K.; Bonas, U. HrpXv, an AraC-type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 1996, 178, 3462–3469. [Google Scholar] [PubMed]
- Zhang, F.; Du, Z.; Huang, L.; Cruz, C.V.; Zhou, Y.; Li, Z. Comparative transcriptome profiling reveals different expression patterns in Xanthomonas oryzae pv. oryzae strains with putative virulence-relevant genes. PLoS ONE 2013, 8, e64267. [Google Scholar]
- Charles, T.C.; Jin, S.; Nester, E.W. Two-component sensory transduction systems in phytobacteria. Annu. Rev. Phytopathol. 1992, 30, 463–484. [Google Scholar] [CrossRef] [PubMed]
- Ng, W.L.; Bassler, B.L. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 2009, 43, 197–222. [Google Scholar] [CrossRef] [PubMed]
- Slater, H.; Alvarez-Morales, A.; Barber, C.E.; Daniels, M.J.; Dow, J.M. A two component system involving an HD-GYP domain protein links cell-cell signaling to pathogenicity gene expression in Xanthomonas campestris. Mol. Microbiol. 2000, 38, 986–1003. [Google Scholar] [CrossRef] [PubMed]
- Ryan, R.P.; Fouhy, Y.; Lucey, J.F.; Crossman, L.C.; Spiro, S.; He, Y.W.; Zhang, L.H.; Heeb, S.; Camara, M.; Williams, P.; et al. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. USA 2006, 103, 6712–6717. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zhang, Y.; Li, J.L.; Wang, N. Diffusible signal factor-mediated quorum sensing plays a central role in coordinating gene expression of Xanthomonas citri subsp citri. Mol. Plant Microbe Interact. 2012, 25, 165–179. [Google Scholar] [CrossRef] [PubMed]
- Ryan, R.P.; McCarthy, Y.; Andrade, M.; Farah, C.S.; Armitage, J.P.; Dow, J.M. Cell–cell signal-dependent dynamic interactions between HD-GYP and GGDEF domain proteins mediate virulence in Xanthomonas campestris. Proc. Natl. Acad. Sci. USA 2010, 107, 5989–5994. [Google Scholar] [CrossRef] [PubMed]
- Song, W.Y.; Wang, G.L.; Chen, L.L.; Kim, H.S.; Pi, L.Y.; Holsten, T.; Gardner, J.; Wang, B.; Zhai, W.X.; Zhu, L.H.; et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 1995, 270, 1804–1806. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.; Han, S.W.; Bartley, L.E.; Ronald, P.C. Unique characteristics of Xanthomonas oryzae pv. oryzae AvrXa21 and implications for plant innate immunity. Proc. Natl. Acad. Sci. USA 2006, 103, 18395–18400. [Google Scholar] [CrossRef] [PubMed]
- Zwir, I.; Shin, D.; Kato, A.; Nishino, K.; Latifi, T.; Solomon, F.; Hare, J.M.; Huang, H.; Groisman, E.A. Dissecting the PhoP regulatory network of Escherchia coli and Salmonella enterica. Proc. Natl. Acad. Sci. USA 2005, 102, 2862–2867. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Calderon, C.B.; Casadesús, J.; Ramos-Morales, F. Rcs and PhoPQ regulatory overlap in the control of Salmonella enterica virulence. J. Bacteriol. 2007, 189, 6635–6644. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.; Jeong, K.S.; Han, S.W.; Lee, S.E.; Phee, B.K.; Hahn, T.R.; Ronald, P. The Xanthomonas oryzae pv. oryzae PhoPQ two-component system is required for AvrXA21 activity, hrpG expression, and virulence. J. Bacteriol. 2008, 190, 2183–2197. [Google Scholar] [PubMed]
- Bahar, O.; Pruitt, R.; Luu, D.D.; Schwessinger, B.; Daudi, A.; Liu, F.; Ruan, R.; Fontaine-Bodin, L.; Koebnik, R.; Ronald, P. The Xanthomonas Ax21 protein is processed by the general secretory system and is secreted in association with outer membrane vesicles. Peer J. 2014, 2, e242. [Google Scholar] [CrossRef] [PubMed]
- Leach, J.E.; White, F.F. Bacterial avirulence genes. Annu. Rev. Phytopathol. 1996, 34, 153–179. [Google Scholar] [CrossRef] [PubMed]
- Leach, J.E.; Davidson, R.; Liu, B.; Manosalva, P.; Mauleon, R.; Carrillo, G.; Bruce, M.; Stephens, J.; Diaz, M.G.; Nelson, R.; et al. Understanding broad-spectrum durable resistance in rice. In Rice Genetics V; Brar, D.S., Mackill, D.J., Hardy, B., Eds.; World Scientific: Toh Tuck, Singapore, 2007; pp. 191–207. [Google Scholar]
- Pugsley, A.P. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 1993, 57, 50–108. [Google Scholar] [PubMed]
- Lindgren, P.B. The role of hrp genes during plant–bacterial interactions. Annu. Rev. Phytopathol. 1997, 35, 129–152. [Google Scholar] [CrossRef] [PubMed]
- He, S.Y. Type III protein secretion systems in plant and animal pathogenic bacteria. Annu. Rev. Phytopathol. 1998, 36, 363–392. [Google Scholar] [CrossRef] [PubMed]
- Lindgren, P.B.; Peet, R.C.; Panopoulos, N.J. Gene-cluster of Pseudomonas syringae pv. “phaseolicola” controls pathogenicity of bean plants and hypersensitivity of nonhost plants. J. Bacteriol. 1986, 168, 512–522. [Google Scholar] [PubMed]
- Cox, M.P.; Peterson, D.A.; Biggs, P.J. SolexaQA: At a glance quality assessment of illumine second-generation sequencing data. BMC Bioinform. 2010. [Google Scholar] [CrossRef] [PubMed]





| Gene | Locus Tag | Product | RPKM | DEGseq Analysis | |||
|---|---|---|---|---|---|---|---|
| 10331 | 1009 | log2 FC | p-Value | Signature * | |||
| hpaF | XOO0065 | protein HpaF | 13.28 | 26.08 | 0.97 | 0.06798 | FALSE |
| hrpF | XOO0066 | HrpF protein | 58.03 | 98.96 | 0.77 | 0.00487 | FALSE |
| hpa4 | XOO0074 | hypothetical protein | 21.87 | 77.43 | 1.82 | 0.00000 | TRUE |
| hpaB | XOO0075 | protein HpaB | 13.95 | 63.98 | 2.20 | 0.00000 | TRUE |
| hrpE | XOO0076 | hypothetical protein | 24.12 | 55.05 | 1.19 | 0.00139 | FALSE |
| hrpD6 | XOO0077 | protein HrpD6 | 23.53 | 55.18 | 1.23 | 0.00099 | TRUE |
| hrpD5 | XOO0078 | protein HrpD5 | 14.07 | 46.30 | 1.72 | 0.00007 | TRUE |
| hpaA | XOO0079 | protein HpaA | 8.59 | 37.25 | 2.12 | 0.00003 | TRUE |
| hrcS | XOO0080 | hypothetical protein | 8.59 | 41.37 | 2.27 | 0.00000 | TRUE |
| hrcR | XOO0081 | type III secretion system protein | 20.31 | 56.68 | 1.48 | 0.00009 | TRUE |
| hrcQ | XOO0082 | hypothetical protein | 12.61 | 38.41 | 1.61 | 0.00058 | TRUE |
| hpaP | XOO0083 | hrpC3 | 6.95 | 29.32 | 2.08 | 0.00027 | TRUE |
| hrcV | XOO0084 | protein HrcV | 6.78 | 23.74 | 1.81 | 0.00303 | FALSE |
| hrcU | XOO0085 | type III secretion system protein HrcU | 5.77 | 17.98 | 1.64 | 0.01677 | FALSE |
| hrpB1 | XOO0086 | protein HrpB1 | 10.00 | 30.18 | 1.59 | 0.00244 | FALSE |
| hrpB2 | XOO0087 | protein HrpB2 | 8.59 | 51.32 | 2.58 | 0.00000 | TRUE |
| hrpB3 (hrcJ) | XOO0088 | protein HrpB3 | 9.03 | 23.62 | 1.39 | 0.01673 | FALSE |
| hrpB4 | XOO0089 | protein HrpB4 | 8.46 | 27.56 | 1.70 | 0.00228 | FALSE |
| hrpB5 | XOO0090 | type III secretion system protein HrpB | 6.72 | 23.82 | 1.83 | 0.00276 | FALSE |
| hrcN | XOO0091 | type III secretion system ATPase | 5.06 | 19.22 | 1.93 | 0.00519 | FALSE |
| hrpB7 | XOO0092 | protein HrpB7 | 5.91 | 22.69 | 1.94 | 0.00225 | FALSE |
| hrpB8 (hrcT) | XOO0093 | protein HrpB8 | 6.60 | 26.50 | 2.00 | 0.00073 | TRUE |
| hrcC | XOO0094 | hypothetical protein | 14.79 | 33.69 | 1.19 | 0.01258 | FALSE |
| hpa1 | XOO0095 | protein Hpa1 | 15.64 | 41.50 | 1.41 | 0.00132 | FALSE |
| hpa2 | XOO0096 | protein Hpa2 | 68.65 | 145.14 | 1.08 | 0.00000 | TRUE |
| hrpG | XOO1379 | HrpG protein | 9.59 | 14.98 | 0.64 | 0.36548 | FALSE |
| hrpXct | XOO1380 | HrpX protein | 8.26 | 8.83 | 0.10 | 0.98273 | FALSE |
| hpa3 | XOO4700 | hypothetical protein | 69.68 | 181.24 | 1.38 | 0.00000 | TRUE |
| Gene | Locus Tag | Product | RPKM | DEGseq Analysis | |||
|---|---|---|---|---|---|---|---|
| 10331 | 1009 | log2 FC | p-Value | Signature * | |||
| ygiY | XOO0057 | two-component system sensor protein | 23.33 | 43.05 | 0.77 | 0.03277 | FALSE |
| – | XOO0336 | two-component system sensor protein | 6.14 | 25.69 | 1.96 | 0.00068 | TRUE |
| phoP | XOO0423 | two-component system regulatory protein | 143.04 | 99.05 | −0.64 | 0.00061 | TRUE |
| phoQ | XOO0424 | two-component system sensor protein | 42.90 | 38.67 | −0.26 | 0.41585 | FALSE |
| – | XOO0519 | two-component system sensor protein | 8.56 | 37.48 | 2.02 | 0.00003 | TRUE |
| – | XOO0520 | two-component system regulatory protein | 10.45 | 27.08 | 1.26 | 0.01107 | FALSE |
| – | XOO0683 | two-component response regulator | 14.10 | 16.94 | 0.15 | 0.76601 | FALSE |
| tctD | XOO1105 | two-component system regulatory protein | 58.18 | 117.85 | 0.91 | 0.00005 | TRUE |
| tctE | XOO1106 | two-component system sensor protein | 21.90 | 52.13 | 1.14 | 0.00115 | FALSE |
| colS | XOO1207 | two-component system sensor protein | 66.31 | 231.53 | 1.69 | 0.00000 | TRUE |
| colR | XOO1208 | two-component system regulatory protein | 26.59 | 48.57 | 0.76 | 0.02578 | FALSE |
| creC | XOO1477 | two-component system sensor protein | 69.24 | 97.83 | 0.39 | 0.08422 | FALSE |
| baeS | XOO1558 | two-component system sensor protein | 3.69 | 13.48 | 1.76 | 0.02205 | FALSE |
| baeR | XOO1559 | two-component system regulatory protein | 5.03 | 12.25 | 1.17 | 0.10729 | FALSE |
| pilR | XOO1591 | two-component system regulatory protein | 21.08 | 18.95 | −0.26 | 0.56344 | FALSE |
| pilS | XOO1592 | two-component system sensor protein | 5.36 | 10.62 | 0.88 | 0.23802 | FALSE |
| regR | XOO2227 | two-component system regulatory protein | 117.22 | 376.62 | 1.57 | 0.00000 | TRUE |
| regS | XOO2228 | two-component system sensor protein | 43.39 | 165.10 | 1.82 | 0.00000 | TRUE |
| – | XOO2322 | two-component system regulatory protein | 124.34 | 48.71 | −1.46 | 0.00000 | TRUE |
| – | XOO2323 | two-component system sensor protein | 16.32 | 17.80 | 0.02 | 0.97542 | FALSE |
| rrpX | XOO2787 | transcriptional regulator | 13.09 | 21.30 | 0.59 | 0.23576 | FALSE |
| – | XOO2797 | two-component system sensor protein | 10.54 | 12.89 | 0.18 | 0.76266 | FALSE |
| – | XOO2798 | two-component system regulatory protein | 32.57 | 7.65 | −2.20 | 0.00002 | TRUE |
| rpfC | XOO2870 | RpfC protein | 18.06 | 27.96 | 0.52 | 0.22665 | FALSE |
| rpfG | XOO2871 | response regulator | 39.38 | 52.65 | 0.31 | 0.30732 | FALSE |
| – | XOO3527 | two-component system regulatory protein | 21.45 | 24.84 | 0.10 | 0.81159 | FALSE |
| – | XOO3528 | two-component system sensor protein | 9.75 | 14.03 | 0.41 | 0.48663 | FALSE |
| – | XOO3659 | two-component system regulatory protein | 114.21 | 209.67 | 0.77 | 0.00000 | TRUE |
| phoB | XOO3666 | two-component system regulatory protein | 11.54 | 18.72 | 0.59 | 0.26937 | FALSE |
| phoR | XOO3667 | two-component system sensor protein | 4.77 | 13.97 | 1.44 | 0.04384 | FALSE |
| torS | XOO3709 | two-component system sensor protein | 7.90 | 34.31 | 2.01 | 0.00006 | TRUE |
| – | XOO3710 | two-component system regulatory protein | 7.34 | 38.26 | 2.27 | 0.00001 | TRUE |
| colS | XOO3762 | two-component system sensor protein | 8.93 | 13.62 | 0.50 | 0.41805 | FALSE |
| colR | XOO3763 | two-component system regulatory protein | 59.32 | 42.52 | −0.59 | 0.04006 | FALSE |
| kdpE | XOO3842 | two-component system regulatory protein | 5.43 | 13.43 | 1.20 | 0.08655 | FALSE |
| kdpD | XOO3843 | two-component system sensor protein | 10.16 | 13.90 | 0.34 | 0.56471 | FALSE |
| – | XOO3870 | two-component system regulatory protein | 35.44 | 60.64 | 0.66 | 0.02655 | FALSE |
| – | XOO3871 | two-component system sensor protein | 14.79 | 43.80 | 1.46 | 0.00032 | TRUE |
| – | XOO3875 | two-component system sensor protein | 164.50 | 381.45 | 1.10 | 0.00000 | TRUE |
| – | XOO3935 | two-component system regulatory protein | 11.04 | 15.52 | 0.38 | 0.49899 | FALSE |
| – | XOO3936 | two-component system sensor protein | 16.87 | 19.88 | 0.13 | 0.79059 | FALSE |
| algR | XOO4008 | two-component system regulatory protein | 26.39 | 59.48 | 1.06 | 0.00105 | FALSE |
| algZ | XOO4009 | two-component system sensor protein | 13.28 | 19.44 | 0.44 | 0.38875 | FALSE |
| – | XOO4201 | two-component system sensor protein | 20.73 | 26.41 | 0.24 | 0.57069 | FALSE |
| ntrC | XOO4202 | two-component system regulatory protein | 8.72 | 15.56 | 0.72 | 0.22512 | FALSE |
| smeR | XOO4341 | two-component system regulatory protein | 7.69 | 18.97 | 1.19 | 0.04211 | FALSE |
| ntrC | XOO4483 | two-component system regulatory protein | 13.97 | 20.70 | 0.46 | 0.35612 | FALSE |
| ntrB | XOO4484 | two-component system sensor protein | 40.25 | 58.94 | 0.44 | 0.13231 | FALSE |
| tcsR | XOO4543 | two-component system regulatory protein | 7.25 | 18.38 | 1.23 | 0.04017 | FALSE |
| raxP | XOO3397 | sulfate adenylyltransferase subunit 2 | 10.46 | 18.71 | 0.73 | 0.18138 | FALSE |
| colS (raxH) | XOO3534 | two-component system sensor protein | 7.85 | 14.60 | 0.78 | 0.20791 | FALSE |
| colS (raxR) | XOO3535 | two-component system regulatory protein | 11.88 | 18.41 | 0.52 | 0.32541 | FALSE |
| raxB | XOO3543 | ABC transporter protein RaxB | 8.04 | 15.86 | 0.87 | 0.15188 | FALSE |
| raxA | XOO3544 | membrane fusion protein RaxA | 3.86 | 12.99 | 1.64 | 0.03244 | FALSE |
| raxST | XOO3546 | sulfotransferase RaxST | 11.06 | 12.49 | 0.07 | 0.91282 | FALSE |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noh, T.-H.; Song, E.-S.; Kim, H.-I.; Kang, M.-H.; Park, Y.-J. Transcriptome-Based Identification of Differently Expressed Genes from Xanthomonas oryzae pv. oryzae Strains Exhibiting Different Virulence in Rice Varieties. Int. J. Mol. Sci. 2016, 17, 259. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17020259
Noh T-H, Song E-S, Kim H-I, Kang M-H, Park Y-J. Transcriptome-Based Identification of Differently Expressed Genes from Xanthomonas oryzae pv. oryzae Strains Exhibiting Different Virulence in Rice Varieties. International Journal of Molecular Sciences. 2016; 17(2):259. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17020259
Chicago/Turabian StyleNoh, Tae-Hwan, Eun-Sung Song, Hong-Il Kim, Mi-Hyung Kang, and Young-Jin Park. 2016. "Transcriptome-Based Identification of Differently Expressed Genes from Xanthomonas oryzae pv. oryzae Strains Exhibiting Different Virulence in Rice Varieties" International Journal of Molecular Sciences 17, no. 2: 259. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17020259