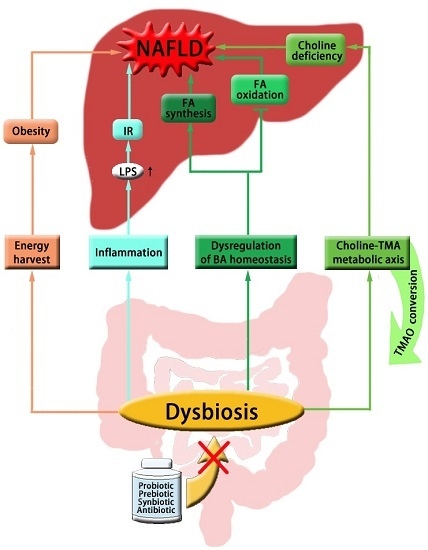
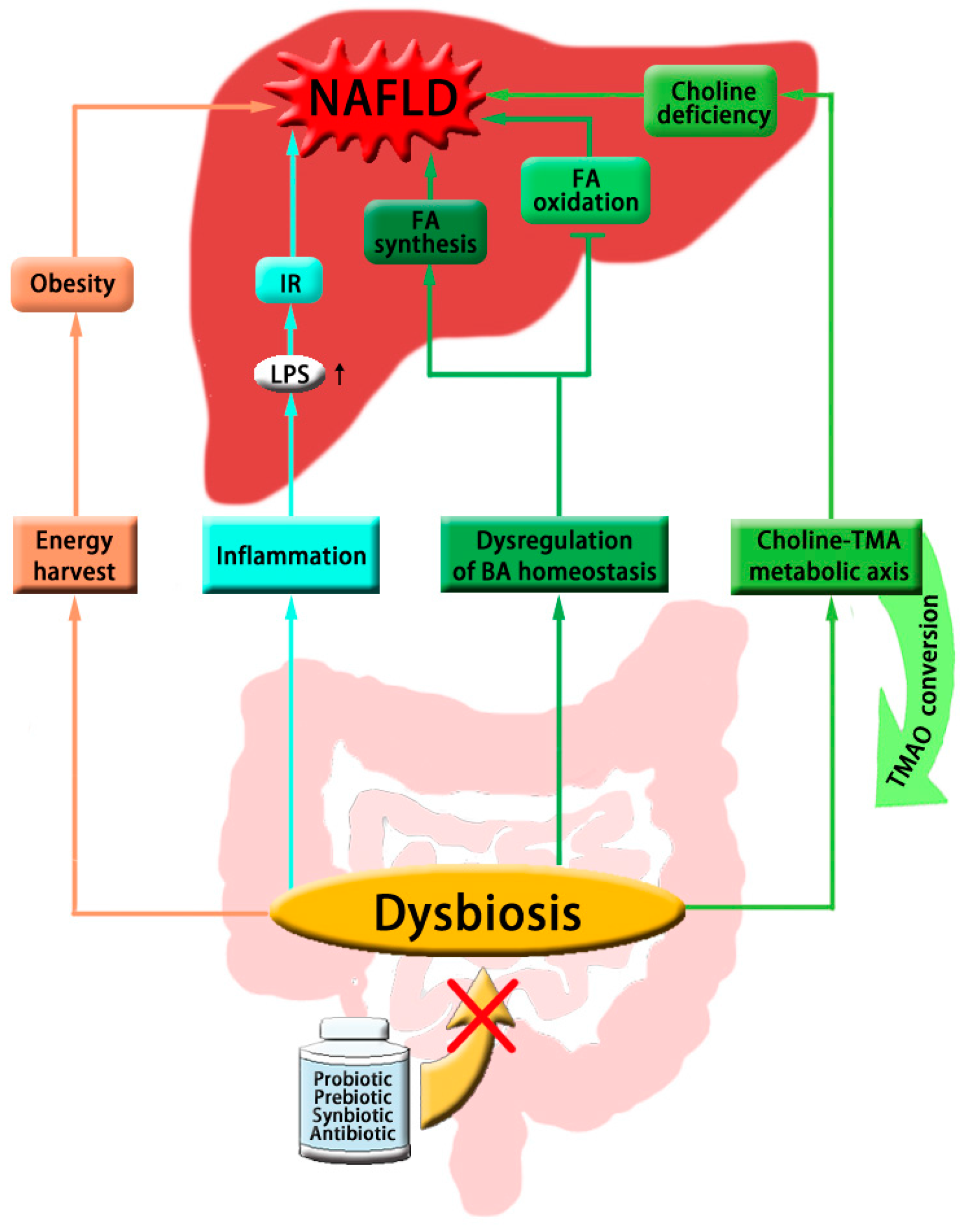
Gut Microbiota and Nonalcoholic Fatty Liver Disease: Insights on Mechanism and Application of Metabolomics
Abstract
:1. Introduction
2. The Gut Microbial Mechanism of NAFLD
2.1. Gut Microbiota and Host Energy Metabolism
2.2. Gut Microbiota and Insulin Sensitivity
2.3. Gut Microbiota and Choline Metabolism
2.4. Gut Microbiota and Bile Acid Metabolism
2.5. Gut Microbiota-Targeted Therapy of NAFLD
3. Application of Metabolomics in Characterizing the Role of Gut Microbiota in NAFLD
3.1. Gut Microbial Metabotypes
3.2. Gut Microbial Metabotypes and NAFLD
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-Gut Microbiota Metabolic Interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [PubMed]
- Hoefert, B. Über die bakterienbefunde im duodenalsaft von gesunden und kranken. Zschr. Klin. Med. 1921, 92, 221–235. (In German) [Google Scholar]
- Abu-Shanab, A.; Quigley, E.M. The role of the gut microbiota in nonalcoholic fatty liver disease. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 691–701. [Google Scholar] [CrossRef] [PubMed]
- Wieland, A.; Frank, D.N.; Harnke, B.; Bambha, K. Systematic review: Microbial dysbiosis and nonalcoholic fatty liver disease. Aliment. Pharmacol. Ther. 2015, 42, 1051–1063. [Google Scholar] [CrossRef] [PubMed]
- Wong, V.W.; Tse, C.H.; Lam, T.T.; Wong, G.L.; Chim, A.M.; Chu, W.C.; Yeung, D.K.; Law, P.T.; Kwan, H.S.; Yu, J.; et al. Molecular characterization of the fecal microbiota in patients with nonalcoholic steatohepatitis—A longitudinal study. PLoS ONE 2013, 8, e62885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Delzenne, N.M.; Cani, P.D.; Everard, A.; Neyrinck, A.M.; Bindels, L.B. Gut microorganisms as promising targets for the management of type 2 diabetes. Diabetologia 2015, 58, 2206–2217. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Obin, M.S.; Zhao, L. The gut microbiota, obesity and insulin resistance. Mol. Asp. Med. 2013, 34, 39–58. [Google Scholar] [CrossRef] [PubMed]
- Escobedo, G.; Lopez-Ortiz, E.; Torres-Castro, I. Gut microbiota as a key player in triggering obesity, systemic inflammation and insulin resistance. Rev. Investig. Clin. 2014, 66, 450–459. [Google Scholar]
- Mehal, W.Z. The Gordian Knot of dysbiosis, obesity and NAFLD. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 637–644. [Google Scholar] [CrossRef] [PubMed]
- Henao-Mejia, J.; Elinav, E.; Jin, C.; Hao, L.; Mehal, W.Z.; Strowig, T.; Thaiss, C.A.; Kau, A.L.; Eisenbarth, S.C.; Jurczak, M.J.; et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 2012, 482, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Wree, A.; Broderick, L.; Canbay, A.; Hoffman, H.M.; Feldstein, A.E. From NAFLD to NASH to cirrhosis-new insights into disease mechanisms. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Mouzaki, M.; Comelli, E.M.; Arendt, B.M.; Bonengel, J.; Fung, S.K.; Fischer, S.E.; McGilvray, I.D.; Allard, J.P. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology 2013, 58, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Le Roy, T.; Llopis, M.; Lepage, P.; Bruneau, A.; Rabot, S.; Bevilacqua, C.; Martin, P.; Philippe, C.; Walker, F.; Bado, A.; et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut 2013, 62, 1787–1794. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Delzenne, N.M. The role of the gut microbiota in energy metabolism and metabolic disease. Curr. Pharm. Des. 2009, 15, 1546–1558. [Google Scholar] [CrossRef] [PubMed]
- Backhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Kalliomaki, M.; Collado, M.C.; Salminen, S.; Isolauri, E. Early differences in fecal microbiota composition in children may predict overweight. Am. J. Clin. Nutr. 2008, 87, 534–538. [Google Scholar] [PubMed]
- Samuel, B.S.; Shaito, A.; Motoike, T.; Rey, F.E.; Backhed, F.; Manchester, J.K.; Hammer, R.E.; Williams, S.C.; Crowley, J.; Yanagisawa, M.; et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc. Natl. Acad. Sci. USA 2008, 105, 16767–16772. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.M.; de Souza, R.; Kendall, C.W.; Emam, A.; Jenkins, D.J. Colonic health: Fermentation and short chain fatty acids. J. Clin. Gastroenterol. 2006, 40, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Tappenden, K.A.; McBurney, M.I. Systemic short-chain fatty acids rapidly alter gastrointestinal structure, function, and expression of early response genes. Dig. Dis. Sci. 1998, 43, 1526–1536. [Google Scholar] [CrossRef] [PubMed]
- Tappenden, K.A.; Drozdowski, L.A.; Thomson, A.B.; McBurney, M.I. Short-chain fatty acid-supplemented total parenteral nutrition alters intestinal structure, glucose transporter 2 (GLUT2) mRNA and protein, and proglucagon mRNA abundance in normal rats. Am. J. Clin. Nutr. 1998, 68, 118–125. [Google Scholar] [PubMed]
- Cani, P.D.; Possemiers, S.; van de Wiele, T.; Guiot, Y.; Everard, A.; Rottier, O.; Geurts, L.; Naslain, D.; Neyrinck, A.; Lambert, D.M.; et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut 2009, 58, 1091–1103. [Google Scholar] [CrossRef] [PubMed]
- Yadav, H.; Lee, J.H.; Lloyd, J.; Walter, P.; Rane, S.G. Beneficial metabolic effects of a probiotic via butyrate-induced GLP-1 hormone secretion. J. Biol. Chem. 2013, 288, 25088–25097. [Google Scholar] [CrossRef] [PubMed]
- Tarantino, G.; Caputi, A. JNKs, insulin resistance and inflammation: A possible link between NAFLD and coronary artery disease. World J. Gastroenterol. 2011, 17, 3785–3794. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Gay, M.A.; de Matias, J.M.; Gonzalez-Juanatey, C.; Garcia-Porrua, C.; Sanchez-Andrade, A.; Martin, J.; Llorca, J. Anti-tumor necrosis factor-α blockade improves insulin resistance in patients with rheumatoid arthritis. Clin. Exp. Rheumatol. 2006, 24, 83–86. [Google Scholar] [PubMed]
- Ma, X.; Hua, J.; Li, Z. Probiotics improve high fat diet-induced hepatic steatosis and insulin resistance by increasing hepatic NKT cells. J. Hepatol. 2008, 49, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Naito, E.; Yoshida, Y.; Makino, K.; Kounoshi, Y.; Kunihiro, S.; Takahashi, R.; Matsuzaki, T.; Miyazaki, K.; Ishikawa, F. Beneficial effect of oral administration of Lactobacillus casei strain Shirota on insulin resistance in diet-induced obesity mice. J. Appl. Microbiol. 2011, 110, 650–657. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yang, S.; Lin, H.; Huang, J.; Watkins, P.A.; Moser, A.B.; Desimone, C.; Song, X.Y.; Diehl, A.M. Probiotics and antibodies to TNF inhibit inflammatory activity and improve nonalcoholic fatty liver disease. Hepatology 2003, 37, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.J.; Membrez, M.; Blancher, F. Gut decontamination with norfloxacin and ampicillin enhances insulin sensitivity in mice. Nestle Nutr. Workshop Ser. Pediatr. Program 2008, 62, 127–137. [Google Scholar] [PubMed]
- Membrez, M.; Blancher, F.; Jaquet, M.; Bibiloni, R.; Cani, P.D.; Burcelin, R.G.; Corthesy, I.; Mace, K.; Chou, C.J. Gut microbiota modulation with norfloxacin and ampicillin enhances glucose tolerance in mice. FASEB J. 2008, 22, 2416–2426. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Bibiloni, R.; Knauf, C.; Waget, A.; Neyrinck, A.M.; Delzenne, N.M.; Burcelin, R. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 2008, 57, 1470–1481. [Google Scholar] [CrossRef] [PubMed]
- Cox, L.M.; Yamanishi, S.; Sohn, J.; Alekseyenko, A.V.; Leung, J.M.; Cho, I.; Kim, S.G.; Li, H.; Gao, Z.; Mahana, D.; et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 2014, 158, 705–721. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.; Park, Y.J.; Kim, Y.R.; Kim, Y.N.; Ka, S.; Lee, H.Y.; Seong, J.K.; Seok, Y.J.; Kim, J.B. Alteration of gut microbiota by vancomycin and bacitracin improves insulin resistance via glucagon-like peptide 1 in diet-induced obesity. FASEB J. 2015, 29, 2397–2411. [Google Scholar] [CrossRef] [PubMed]
- Corbin, K.D.; Zeisel, S.H. Choline metabolism provides novel insights into nonalcoholic fatty liver disease and its progression. Curr. Opin. Gastroenterol. 2012, 28, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Zeisel, S.H. Choline: Critical role during fetal development and dietary requirements in adults. Annu. Rev. Nutr. 2006, 26, 229–250. [Google Scholar] [CrossRef] [PubMed]
- Hebbard, L.; George, J. Animal models of nonalcoholic fatty liver disease. Nat. Rev. Gastroenterol. Hepatol. 2011, 8, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Waite, K.A.; Cabilio, N.R.; Vance, D.E. Choline deficiency-induced liver damage is reversible in Pemt-/- mice. J. Nutr. 2002, 132, 68–71. [Google Scholar] [PubMed]
- Cano, A.; Buque, X.; Martinez-Una, M.; Aurrekoetxea, I.; Menor, A.; Garcia-Rodriguez, J.L.; Lu, S.C.; Martinez-Chantar, M.L.; Mato, J.M.; Ochoa, B.; et al. Methionine adenosyltransferase 1A gene deletion disrupts hepatic very low-density lipoprotein assembly in mice. Hepatology 2011, 54, 1975–1986. [Google Scholar] [CrossRef] [PubMed]
- Zeisel, S.H.; Wishnok, J.S.; Blusztajn, J.K. Formation of methylamines from ingested choline and lecithin. J. Pharmacol. Exp. Ther. 1983, 225, 320–324. [Google Scholar] [PubMed]
- Al-Waiz, M.; Mikov, M.; Mitchell, S.C.; Smith, R.L. The exogenous origin of trimethylamine in the mouse. Metabolism 1992, 41, 135–136. [Google Scholar] [CrossRef]
- Spencer, M.D.; Hamp, T.J.; Reid, R.W.; Fischer, L.M.; Zeisel, S.H.; Fodor, A.A. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology 2011, 140, 976–986. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, C.; Claudel, T.; Trauner, M. Bile acid-mediated control of liver triglycerides. Semin. Liver Dis. 2013, 33, 330–342. [Google Scholar] [CrossRef] [PubMed]
- Houten, S.M.; Watanabe, M.; Auwerx, J. Endocrine functions of bile acids. EMBO J. 2006, 25, 1419–1425. [Google Scholar] [CrossRef] [PubMed]
- Hylemon, P.B.; Zhou, H.P.; Pandak, W.M.; Ren, S.L.; Gil, G.; Dent, P. Bile acids as regulatory molecules. J. Lipid Res. 2009, 50, 1509–1520. [Google Scholar] [CrossRef] [PubMed]
- Claudel, T.; Staels, B.; Kuipers, F. The Farnesoid X receptor—A molecular link between bile acid and lipid and glucose metabolism. Arterioscler. Thromb. Vasc. 2005, 25, 2020–2031. [Google Scholar] [CrossRef] [PubMed]
- Stacey, M.; Webb, M. Studies on the antibacterial properties of the bile acids and some compounds derived from cholanic acid. Proc. R. Soc. Med. 1947, 134, 523–537. [Google Scholar] [CrossRef] [PubMed]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef] [PubMed]
- Russell, D.W. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 2003, 72, 137–174. [Google Scholar] [CrossRef] [PubMed]
- Yokota, A.; Fukiya, S.; Islam, K.B.; Ooka, T.; Ogura, Y.; Hayashi, T.; Hagio, M.; Ishizuka, S. Is bile acid a determinant of the gut microbiota on a high-fat diet? Gut Microbes 2012, 3, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Serino, M.; Luche, E.; Gres, S.; Baylac, A.; Berge, M.; Cenac, C.; Waget, A.; Klopp, P.; Iacovoni, J.; Klopp, C.; et al. Metabolic adaptation to a high-fat diet is associated with a change in the gut microbiota. Gut 2011, 61, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.; Ahn, S.H.; Inagaki, T.; Choi, M.; Ito, S.; Guo, G.L.; Kliewer, S.A.; Gonzalez, F.J. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J. Lipid Res. 2007, 48, 2664–2672. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, T.; Li, F.; Gonzalez, F.J. FXR signaling in the enterohepatic system. Mol. Cell. Endocrinol. 2013, 368, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Sayin, S.I.; Wahlstrom, A.; Felin, J.; Jantti, S.; Marschall, H.U.; Bamberg, K.; Angelin, B.; Hyotylainen, T.; Oresic, M.; Backhed, F. Gut Microbiota Regulates Bile Acid Metabolism by Reducing the Levels of Tauro-β-muricholic Acid, a Naturally Occurring FXR Antagonist. Cell Metab. 2013, 17, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Jiang, C.T.; Krausz, K.W.; Li, Y.F.; Albert, I.; Hao, H.P.; Fabre, K.M.; Mitchell, J.B.; Patterson, A.D.; Gonzalez, F.J. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Aron-Wisnewsky, J.; Gaborit, B.; Dutour, A.; Clement, K. Gut microbiota and non-alcoholic fatty liver disease: New insights. Clin. Microbiol. Infect. 2013, 19, 338–348. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Xie, C.; Li, F.; Zhang, L.; Nichols, R.G.; Krausz, K.W.; Cai, J.; Qi, Y.; Fang, Z.Z.; Takahashi, S.; et al. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J. Clin. Investig. 2015, 125, 386–402. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lee, F.Y.; Barrera, G.; Lee, H.; Vales, C.; Gonzalez, F.J.; Willson, T.M.; Edwards, P.A. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc. Natl. Acad. Sci. USA 2006, 103, 1006–1011. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Saha, P.K.; Chan, L.; Moore, D.D. Farnesoid X receptor is essential for normal glucose homeostasis. J. Clin. Investig. 2006, 116, 1102–1109. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Horai, Y.; Houten, S.M.; Morimoto, K.; Sugizaki, T.; Arita, E.; Mataki, C.; Sato, H.; Tanigawara, Y.; Schoonjans, K.; et al. Lowering bile acid pool size with a synthetic farnesoid X receptor (FXR) agonist induces obesity and diabetes through reduced energy expenditure. J. Biol. Chem. 2011, 286, 26913–26920. [Google Scholar] [CrossRef] [PubMed]
- Hirokane, H.; Nakahara, M.; Tachibana, S.; Shimizu, M.; Sato, R. Bile acid reduces the secretion of very low density lipoprotein by repressing microsomal triglyceride transfer protein gene expression mediated by hepatocyte nuclear factor-4. J. Biol. Chem. 2004, 279, 45685–45692. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Li, F.; Guo, G.L. Tissue-specific function of farnesoid X receptor in liver and intestine. Pharmacol. Res. 2011, 63, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Anakk, S.; Watanabe, M.; Ochsner, S.A.; McKenna, N.J.; Finegold, M.J.; Moore, D.D. Combined deletion of Fxr and Shp in mice induces Cyp17a1 and results in juvenile onset cholestasis. J. Clin. Investig. 2011, 121, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Urizar, N.L.; Liverman, A.B.; Dodds, D.T.; Silva, F.V.; Ordentlich, P.; Yan, Y.; Gonzalez, F.J.; Heyman, R.A.; Mangelsdorf, D.J.; Moore, D.D. A natural product that lowers cholesterol as an antagonist ligand for FXR. Science 2002, 296, 1703–1706. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Huang, L.; Zhao, A.; Lew, J.L.; Yu, J.; Sahoo, S.; Meinke, P.T.; Royo, I.; Pelaez, F.; Wright, S.D. Guggulsterone is a farnesoid X receptor antagonist in coactivator association assays but acts to enhance transcription of bile salt export pump. J. Biol. Chem. 2003, 278, 10214–10220. [Google Scholar] [CrossRef] [PubMed]
- Nabavi, S.; Rafraf, M.; Somi, M.H.; Homayouni-Rad, A.; Asghari-Jafarabadi, M. Effects of probiotic yogurt consumption on metabolic factors in individuals with nonalcoholic fatty liver disease. J. Dairy Sci. 2014, 97, 7386–7393. [Google Scholar] [CrossRef] [PubMed]
- Aller, R.; De Luis, D.A.; Izaola, O.; Conde, R.; Gonzalez Sagrado, M.; Primo, D.; de La Fuente, B.; Gonzalez, J. Effect of a probiotic on liver aminotransferases in nonalcoholic fatty liver disease patients: A double blind randomized clinical trial. Eur. Rev. Med. Pharmacol. Sci. 2011, 15, 1090–1095. [Google Scholar] [PubMed]
- Ren, T.; Huang, C.; Cheng, M. Dietary blueberry and bifidobacteria attenuate nonalcoholic fatty liver disease in rats by affecting SIRT1-mediated signaling pathway. Oxid. Med. Cell. Longev. 2014, 2014, 469059. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.Y.; Wan, Y.P.; Fang, Q.Y.; Lu, W.; Cai, W. Supplementation with probiotics modifies gut flora and attenuates liver fat accumulation in rat nonalcoholic fatty liver disease model. J. Clin. Biochem. Nutr. 2012, 50, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tang, H.; Zhang, C.; Zhao, Y.; Derrien, M.; Rocher, E.; van-Hylckama Vlieg, J.E.; Strissel, K.; Zhao, L.; Obin, M.; et al. Modulation of gut microbiota during probiotic-mediated attenuation of metabolic syndrome in high fat diet-fed mice. ISME J. 2015, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Plaza-Diaz, J.; Gomez-Llorente, C.; Abadia-Molina, F.; Saez-Lara, M.J.; Campana-Martin, L.; Munoz-Quezada, S.; Romero, F.; Gil, A.; Fontana, L. Effects of Lactobacillus paracasei CNCM I-4034, Bifidobacterium breve CNCM I-4035 and Lactobacillus rhamnosus CNCM I-4036 on hepatic steatosis in Zucker rats. PLoS ONE 2014, 9, e98401. [Google Scholar] [CrossRef]
- Loguercio, C.; Federico, A.; Tuccillo, C.; Terracciano, F.; D’Auria, M.V.; De Simone, C.; del Vecchio Blanco, C. Beneficial effects of a probiotic VSL#3 on parameters of liver dysfunction in chronic liver diseases. J. Clin. Gastroenterol. 2005, 39, 540–543. [Google Scholar] [PubMed]
- Vajro, P.; Mandato, C.; Licenziati, M.R.; Franzese, A.; Vitale, D.F.; Lenta, S.; Caropreso, M.; Vallone, G.; Meli, R. Effects of Lactobacillus rhamnosus strain GG in pediatric obesity-related liver disease. J. Pediatr. Gastroenterol. Nutr. 2011, 52, 740–743. [Google Scholar] [CrossRef] [PubMed]
- Cesaro, C.; Tiso, A.; Del Prete, A.; Cariello, R.; Tuccillo, C.; Cotticelli, G.; del Vecchio Blanco, C.; Loguercio, C. Gut microbiota and probiotics in chronic liver diseases. Dig. Liver Dis. 2011, 43, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Frazier, T.H.; DiBaise, J.K.; McClain, C.J. Gut microbiota, intestinal permeability, obesity-induced inflammation, and liver injury. JPEN J. Parenter. Enteral Nutr. 2011, 35, 14S–20S. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, E.F.; Cotter, P.D.; Stanton, C.; Ross, R.P.; Hill, C. Production of bioactive substances by intestinal bacteria as a basis for explaining probiotic mechanisms: Bacteriocins and conjugated linoleic acid. Int. J. Food Microbiol. 2012, 152, 189–205. [Google Scholar] [CrossRef] [PubMed]
- Ferolla, S.M.; Armiliato, G.N.; Couto, C.A.; Ferrari, T.C. Probiotics as a complementary therapeutic approach in nonalcoholic fatty liver disease. World J. Hepatol. 2015, 7, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Iacono, A.; Raso, G.M.; Canani, R.B.; Calignano, A.; Meli, R. Probiotics as an emerging therapeutic strategy to treat NAFLD: Focus on molecular and biochemical mechanisms. J. Nutr. Biochem. 2011, 22, 699–711. [Google Scholar] [CrossRef] [PubMed]
- Flint, H.J. Obesity and the gut microbiota. J. Clin. Gastroenterol. 2011, 45, S128–S132. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.H.; Nishimura, Y.; Hishikawa, D.; Tsuzuki, H.; Miyahara, H.; Gotoh, C.; Choi, K.C.; Feng, D.D.; Chen, C.; Lee, H.G.; et al. Acetate and propionate short chain fatty acids stimulate adipogenesis via GPCR43. Endocrinology 2005, 146, 5092–5099. [Google Scholar] [CrossRef] [PubMed]
- Layden, B.T.; Angueira, A.R.; Brodsky, M.; Durai, V.; Lowe, W.L., Jr. Short chain fatty acids and their receptors: New metabolic targets. Transl. Res. 2013, 161, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Sleeth, M.L.; Thompson, E.L.; Ford, H.E.; Zac-Varghese, S.E.; Frost, G. Free fatty acid receptor 2 and nutrient sensing: A proposed role for fibre, fermentable carbohydrates and short-chain fatty acids in appetite regulation. Nutr. Res. Rev. 2010, 23, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Maurer, A.D.; Eller, L.K.; Hallam, M.C.; Taylor, K.; Reimer, R.A. Consumption of diets high in prebiotic fiber or protein during growth influences the response to a high fat and sucrose diet in adulthood in rats. Nutr. Metab. 2010, 7, 77. [Google Scholar] [CrossRef] [PubMed]
- Pyra, K.A.; Saha, D.C.; Reimer, R.A. Prebiotic fiber increases hepatic acetyl CoA carboxylase phosphorylation and suppresses glucose-dependent insulinotropic polypeptide secretion more effectively when used with metformin in obese rats. J. Nutr. 2012, 142, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Delzenne, N.M.; Neyrinck, A.M.; Cani, P.D. Modulation of the gut microbiota by nutrients with prebiotic properties: Consequences for host health in the context of obesity and metabolic syndrome. Microb. Cell Fact. 2011, 10 (Suppl. S1), S10. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Delzenne, N.M. The gut microbiome as therapeutic target. Pharmacol. Ther. 2011, 130, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Malaguarnera, M.; Vacante, M.; Antic, T.; Giordano, M.; Chisari, G.; Acquaviva, R.; Mastrojeni, S.; Malaguarnera, G.; Mistretta, A.; Li Volti, G.; et al. Bifidobacterium longum with fructo-oligosaccharides in patients with non alcoholic steatohepatitis. Dig. Dis. Sci. 2012, 57, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Wong, V.W.; Won, G.L.; Chim, A.M.; Chu, W.C.; Yeung, D.K.; Li, K.C.; Chan, H.L. Treatment of nonalcoholic steatohepatitis with probiotics. A proof-of-concept study. Ann. Hepatol. 2013, 12, 256–262. [Google Scholar] [PubMed]
- Xie, G.; Zhang, S.; Zheng, X.; Jia, W. Metabolomics approaches for characterizing metabolic interactions between host and its commensal microbes. Electrophoresis 2013, 34, 2787–2798. [Google Scholar] [CrossRef] [PubMed]
- Everett, J.R. Pharmacometabonomics in humans: A new tool for personalized medicine. Pharmacogenomics 2015, 16, 737–754. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Xie, G.; Zhao, A.; Zhao, L.; Yao, C.; Chiu, N.H.; Zhou, Z.; Bao, Y.; Jia, W.; Nicholson, J.K.; et al. The footprints of gut microbial-mammalian co-metabolism. J. Proteome Res. 2011, 10, 5512–5522. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.P.; Dumas, M.E.; Wang, Y.; Legido-Quigley, C.; Yap, I.K.; Tang, H.; Zirah, S.; Murphy, G.M.; Cloarec, O.; Lindon, J.C.; et al. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol. Syst. Biol. 2007, 3, 112. [Google Scholar] [CrossRef] [PubMed]
- Dumas, M.E.; Barton, R.H.; Toye, A.; Cloarec, O.; Blancher, C.; Rothwell, A.; Fearnside, J.; Tatoud, R.; Blanc, V.; Lindon, J.C.; et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc. Natl. Acad. Sci. USA 2006, 103, 12511–12516. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.; Huo, T.I.; Cheng, H.Y.; Tsai, J.C.; Liao, J.W.; Lee, M.S.; Qin, X.M.; Hsieh, M.T.; Pao, L.H.; Peng, W.H. Gallic acid ameliorated impaired glucose and lipid homeostasis in high fat diet-induced NAFLD mice. PLoS ONE 2014, 9, e96969. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, Y.; Zhang, M.; Pang, X.; Xu, J.; Kang, C.; Li, M.; Zhang, C.; Zhang, Z.; Zhang, Y.; et al. Structural changes of gut microbiota during berberine-mediated prevention of obesity and insulin resistance in high-fat diet-fed rats. PLoS ONE 2012, 7, e42529. [Google Scholar] [CrossRef] [PubMed]
- Roelofsen, H.; Priebe, M.G.; Vonk, R.J. The interaction of short-chain fatty acids with adipose tissue: Relevance for prevention of type 2 diabetes. Benef. Microbes 2010, 1, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Baker, R.D.; Baker, S.S. Gut microbiome and nonalcoholic fatty liver diseases. Pediatr. Res. 2015, 77, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Kalhan, S.C.; Guo, L.; Edmison, J.; Dasarathy, S.; McCullough, A.J.; Hanson, R.W.; Milburn, M. Plasma metabolomic profile in nonalcoholic fatty liver disease. Metabolism 2011, 60, 404–413. [Google Scholar] [CrossRef] [PubMed]
- Encarnacao, J.C.; Abrantes, A.M.; Pires, A.S.; Botelho, M.F. Revisit dietary fiber on colorectal cancer: Butyrate and its role on prevention and treatment. Cancer Metastasis Rev. 2015, 34, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Jena, G. The role of butyrate, a histone deacetylase inhibitor in diabetes mellitus: Experimental evidence for therapeutic intervention. Epigenomics 2015, 7, 669–680. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Dong, L.; Xu, W.; Bai, K.; Lu, C.; Wu, Y.; Huang, Q.; Zhang, L.; Wang, T. Dietary Tributyrin Supplementation Attenuates Insulin Resistance and Abnormal Lipid Metabolism in Suckling Piglets with Intrauterine Growth Retardation. PLoS ONE 2015, 10, e0136848. [Google Scholar] [CrossRef] [PubMed]
- Demigne, C.; Morand, C.; Levrat, M.A.; Besson, C.; Moundras, C.; Remesy, C. Effect of propionate on fatty acid and cholesterol synthesis and on acetate metabolism in isolated rat hepatocytes. Br. J. Nutr. 1995, 74, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Katona, M.; Hegyi, P.; Kui, B.; Balla, Z.; Rakonczay, Z., Jr.; Razga, Z.; Tiszlavicz, L.; Maleth, J.; Venglovecz, V. A novel, protective role of ursodeoxycholate in bile-induced pancreatic ductal injury. Am. J. Physiol. Gastrointest. Liver Physiol. 2016, 310, G193–G204. [Google Scholar] [CrossRef] [PubMed]
- Grattagliano, I.; Palmieri, V.O.; Portincasa, P.; Minerva, F.; Palasciano, G. Long-term ursodeoxycholate improves circulating redox changes in primary biliary cirrhotic patients. Clin. Biochem. 2011, 44, 1400–1404. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Lee, Y.K.; Kim, J.W.; Baik, S.K.; Kwon, S.O.; Jang, H.I. Modulation of colon cancer cell invasiveness induced by deoxycholic acid. Korean J. Gastroenterol. 2006, 48, 9–18. [Google Scholar] [PubMed]
- Yoshimoto, S.; Loo, T.M.; Atarashi, K.; Kanda, H.; Sato, S.; Oyadomari, S.; Iwakura, Y.; Oshima, K.; Morita, H.; Hattori, M.; et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 2013, 499, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Bai, P.S.; Sun, H.; Nan, K.J.; Chen, N.Z.; Qi, X.G. The deoxycholic acid targets miRNA-dependent CAC1 gene expression in multidrug resistance of human colorectal cancer. Int. J. Biochem. Cell Biol. 2012, 44, 2321–2332. [Google Scholar] [CrossRef] [PubMed]
- Ufnal, M.; Zadlo, A.; Ostaszewski, R. TMAO: A small molecule of great expectations. Nutrition 2015, 31, 1317–1323. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Wang, Q.; Li, L. A genome-wide systems analysis reveals strong link between colorectal cancer and trimethylamine N-oxide (TMAO), a gut microbial metabolite of dietary meat and fat. BMC Genom. 2015, 16 (Suppl. 7), S4. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Klipfell, E.; Bennett, B.J.; Koeth, R.; Levison, B.S.; Dugar, B.; Feldstein, A.E.; Britt, E.B.; Fu, X.; Chung, Y.M.; et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011, 472, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.H.; Wang, Z.; Kennedy, D.J.; Wu, Y.; Buffa, J.A.; Agatisa-Boyle, B.; Li, X.S.; Levison, B.S.; Hazen, S.L. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ. Res. 2015, 116, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Lever, M.; George, P.M.; Slow, S.; Bellamy, D.; Young, J.M.; Ho, M.; McEntyre, C.J.; Elmslie, J.L.; Atkinson, W.; Molyneux, S.L.; et al. Betaine and Trimethylamine-N-Oxide as Predictors of Cardiovascular Outcomes Show Different Patterns in Diabetes Mellitus: An Observational Study. PLoS ONE 2014, 9, e114969. [Google Scholar] [CrossRef] [PubMed]
- Ge, C.X.; Yu, R.; Xu, M.X.; Li, P.Q.; Fan, C.Y.; Li, J.M.; Kong, L.D. Betaine prevented fructose-induced NAFLD by regulating LXRα/PPARα pathway and alleviating ER stress in rats. Eur. J. Pharmacol. 2016, 770, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Bohus, E.; Coen, M.; Keun, H.C.; Ebbels, T.M.; Beckonert, O.; Lindon, J.C.; Holmes, E.; Noszal, B.; Nicholson, J.K. Temporal metabonomic modeling of l-arginine-induced exocrine pancreatitis. J. Proteome Res. 2008, 7, 4435–4445. [Google Scholar] [CrossRef] [PubMed]
- Clayton, T.A.; Baker, D.; Lindon, J.C.; Everett, J.R.; Nicholson, J.K. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl. Acad. Sci. USA 2009, 106, 14728–14733. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Fritsche, J.; Wang, J.; Chen, J.; Rittig, K.; Schmitt-Kopplin, P.; Fritsche, A.; Haring, H.U.; Schleicher, E.D.; Xu, G.; et al. Metabonomic fingerprints of fasting plasma and spot urine reveal human pre-diabetic metabolic traits. Metabolomics 2010, 6, 362–374. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.; Jung, Y.; Bang, E.J.; Cho, S.I.; Jang, Y.J.; Kwak, J.M.; Ryu do, H.; Park, S.; Hwang, G.S. Noninvasive diagnosis and evaluation of curative surgery for gastric cancer by using NMR-based metabolomic profiling. Ann. Surg. Oncol. 2014, 21 (Suppl. S4), S736–S742. [Google Scholar] [CrossRef] [PubMed]
- Huo, T.; Xiong, Z.; Lu, X.; Cai, S. Metabonomic study of biochemical changes in urinary of type 2 diabetes mellitus patients after the treatment of sulfonylurea antidiabetic drugs based on ultra-performance liquid chromatography/mass spectrometry. Biomed. Chromatogr. 2015, 29, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Deguchi, T.; Isozaki, K.; Yousuke, K.; Terasaki, T.; Otagiri, M. Involvement of organic anion transporters in the efflux of uremic toxins across the blood-brain barrier. J. Neurochem. 2006, 96, 1051–1059. [Google Scholar] [CrossRef] [PubMed]
- Byrd, D.J.; Berthold, H.W.; Trefz, K.F.; Kochen, W.; Gilli, G.; Scharer, K.; Schuler, H.W.; Asbach, H.W. Indolic tryptophan metabolism in uraemia. Proc. Eur. Dial. Transplant. Assoc. 1976, 12, 347–354. [Google Scholar] [PubMed]
- Keszthelyi, D.; Troost, F.J.; Masclee, A.A. Understanding the role of tryptophan and serotonin metabolism in gastrointestinal function. Neurogastroenterol. Motil. 2009, 21, 1239–1249. [Google Scholar] [CrossRef] [PubMed]
- Yeo, A.; Boyd, P.; Lumsden, S.; Saunders, T.; Handley, A.; Stubbins, M.; Knaggs, A.; Asquith, S.; Taylor, I.; Bahari, B.; et al. Association between a functional polymorphism in the serotonin transporter gene and diarrhoea predominant irritable bowel syndrome in women. Gut 2004, 53, 1452–1458. [Google Scholar] [CrossRef] [PubMed]
- O’Mahony, S.M.; Clarke, G.; Borre, Y.E.; Dinan, T.G.; Cryan, J.F. Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behav. Brain Res. 2015, 277, 32–48. [Google Scholar] [CrossRef] [PubMed]
- Favero, G.; Stacchiotti, A.; Castrezzati, S.; Bonomini, F.; Albanese, M.; Rezzani, R.; Rodella, L.F. Melatonin reduces obesity and restores adipokine patterns and metabolism in obese (ob/ob) mice. Nutr. Res. 2015, 35, 891–900. [Google Scholar] [CrossRef] [PubMed]
- Nduhirabandi, F.; du Toit, E.F.; Lochner, A. Melatonin and the metabolic syndrome: A tool for effective therapy in obesity-associated abnormalities? Acta Physiol. 2012, 205, 209–223. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.; Benno, Y. The relationship between microbiota and polyamine concentration in the human intestine: A pilot study. Microbiol. Immunol. 2007, 51, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, R.; Asim, M.; Hoge, S.; Lewis, N.D.; Singh, K.; Barry, D.P.; de Sablet, T.; Piazuelo, M.B.; Sarvaria, A.R.; Cheng, Y.; et al. Polyamines Impair Immunity to Helicobacter pylori by Inhibiting l-Arginine Uptake Required for Nitric Oxide Production. Gastroenterology 2010, 139, 1686–1698. [Google Scholar] [CrossRef] [PubMed]
- Codoner-Franch, P.; Tavarez-Alonso, S.; Murria-Estal, R.; Herrera-Martin, G.; Alonso-Iglesias, E. Polyamines are increased in obese children and are related to markers of oxidative/nitrosative stress and angiogenesis. J. Clin. Endocrinol. Metab. 2011, 96, 2821–2825. [Google Scholar] [CrossRef] [PubMed]
- Hersoug, L.G.; Moller, P.; Loft, S. Gut microbiota-derived lipopolysaccharide uptake and trafficking to adipose tissue: Implications for inflammation and obesity. Obes. Rev. 2015. [Google Scholar] [CrossRef] [PubMed]
- Jayashree, B.; Bibin, Y.S.; Prabhu, D.; Shanthirani, C.S.; Gokulakrishnan, K.; Lakshmi, B.S.; Mohan, V.; Balasubramanyam, M. Increased circulatory levels of lipopolysaccharide (LPS) and zonulin signify novel biomarkers of proinflammation in patients with type 2 diabetes. Mol. Cell. Biochem. 2014, 388, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Klavins, K.; Koal, T.; Dallmann, G.; Marksteiner, J.; Kemmler, G.; Humpel, C. The ratio of phosphatidylcholines to lysophosphatidylcholines in plasma differentiates healthy controls from patients with Alzheimer’s disease and mild cognitive impairment. Alzheimers Dement. 2015, 1, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Mir, S.A.; Rajagopalan, P.; Jain, A.P.; Khan, A.A.; Datta, K.K.; Mohan, S.V.; Lateef, S.S.; Sahasrabuddhe, N.; Somani, B.L.; Keshava Prasad, T.S.; et al. LC-MS-based serum metabolomic analysis reveals dysregulation of phosphatidylcholines in esophageal squamous cell carcinoma. J. Proteom. 2015, 127, 96–102. [Google Scholar] [CrossRef] [PubMed]
| Type of Metabolites | Representative Microbial Metabolites | Biological Functions | Implicated Diseases | Ref. |
|---|---|---|---|---|
| Short chain fatty acids | Butyrate, acetate, propionate | Anti-inflammatory, induce apoptosis and suppress histone deacetylase; provide energy for gastrointestinal tract epithelial cells; inhibit fatty acid and cholesterol synthesis | Colorectal cancer, diabetes mellitus, insulin resistance and lipid disorders | [21,101,102,103,104] |
| Bile acids | UDCA | Suppresses bile-induced pancreatic ductal injury by reducing apoptosis and mitochondrial damage; inhibits DCA-induced apoptosis and its signaling pathway in oesophageal cancer cells | Cancer, gallstone disease, primary biliary cirrhosis | [105,106] |
| DCA | Involved in senescence-associated secretory phenotype that facilitates hepatocellular carcinoma development in mice; contributes to the development of colorectal cancer by decreasing miR-199a-5p expression, or upregulating invasive and angiogenic potentials of human colon cancer cells | Hepatocellular carcinoma and colorectal cancer | [107,108,109] | |
| Choline | TMAO, TMA, DMA | Elevated plasma levels of TMAO positively correlated with increased risk of major adverse cardiovascular events and colorectal cancer; impairs reverse cholesterol transportation; contributes to renal fibrosis and dysfunction in animals | Cardiovascular diseases, cancers, kidney disease | [110,111,112,113] |
| betaine | Regulates the LXRα/PPARα pathway and alleviates ER stress; predictor of cardiovascular outcomes | NAFLD, diabetes | [114,115] | |
| Phenyl derivatives | hippuric acid, hydroxyhippuric acid, benzoic acid, p-cresol, phenylacetylglycine, phenylacetylglutamine | Biomarkers for the involvement and variation of gut microbiota in diseases; detoxification of xenobiotics | Obesity, metabolic diseases, cancers, pancreatitis | [116,117,118,119,120] |
| Indole derivatives | N-acetyltryptophan, indoleacetate | Indicates the defect in tryptophan metabolism; uremic toxins; | Chronic renal failure | [121,122] |
| serotonin, melatonin | Involved in brain-gut axis function; reduce inflammatory infiltration, regulate adipokine secretion and energy expenditure, improve insulin sensitivity | Anxiety, depression, gastrointestinal dysfunction, obesity, metabolic syndrome | [123,124,125,126,127] | |
| Polyamines | putrescine, spermidine, spermine | Stabilize DNA, RNA and protein molecules, regulate cell proliferation and differentiation; biomarkers of human diseases | Cancer, stroke, renal failure, obesity and diabetes | [128,129,130] |
| Lipids | LPS | Impairs intestinal permeability, induces systemic inflammation and insulin resistance | obesity, diabetes | [131,132] |
| phosphatidylcholines | Altered in patients with Alzheimer’s disease or esophageal squamous cell carcinoma | Alzheimer’s disease, patients with esophageal squamous cell carcinoma | [133,134] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, X.; Ji, G.; Jia, W.; Li, H. Gut Microbiota and Nonalcoholic Fatty Liver Disease: Insights on Mechanism and Application of Metabolomics. Int. J. Mol. Sci. 2016, 17, 300. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17030300
He X, Ji G, Jia W, Li H. Gut Microbiota and Nonalcoholic Fatty Liver Disease: Insights on Mechanism and Application of Metabolomics. International Journal of Molecular Sciences. 2016; 17(3):300. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17030300
Chicago/Turabian StyleHe, Xuyun, Guang Ji, Wei Jia, and Houkai Li. 2016. "Gut Microbiota and Nonalcoholic Fatty Liver Disease: Insights on Mechanism and Application of Metabolomics" International Journal of Molecular Sciences 17, no. 3: 300. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17030300