Molecular Characterization and Expression Analyses of the Complement Component C8α, C8β and C9 Genes in Yellow Catfish (Pelteobagrus fulvidraco) after the Aeromonas hydrophila Challenge
Abstract
:1. Introduction
2. Results
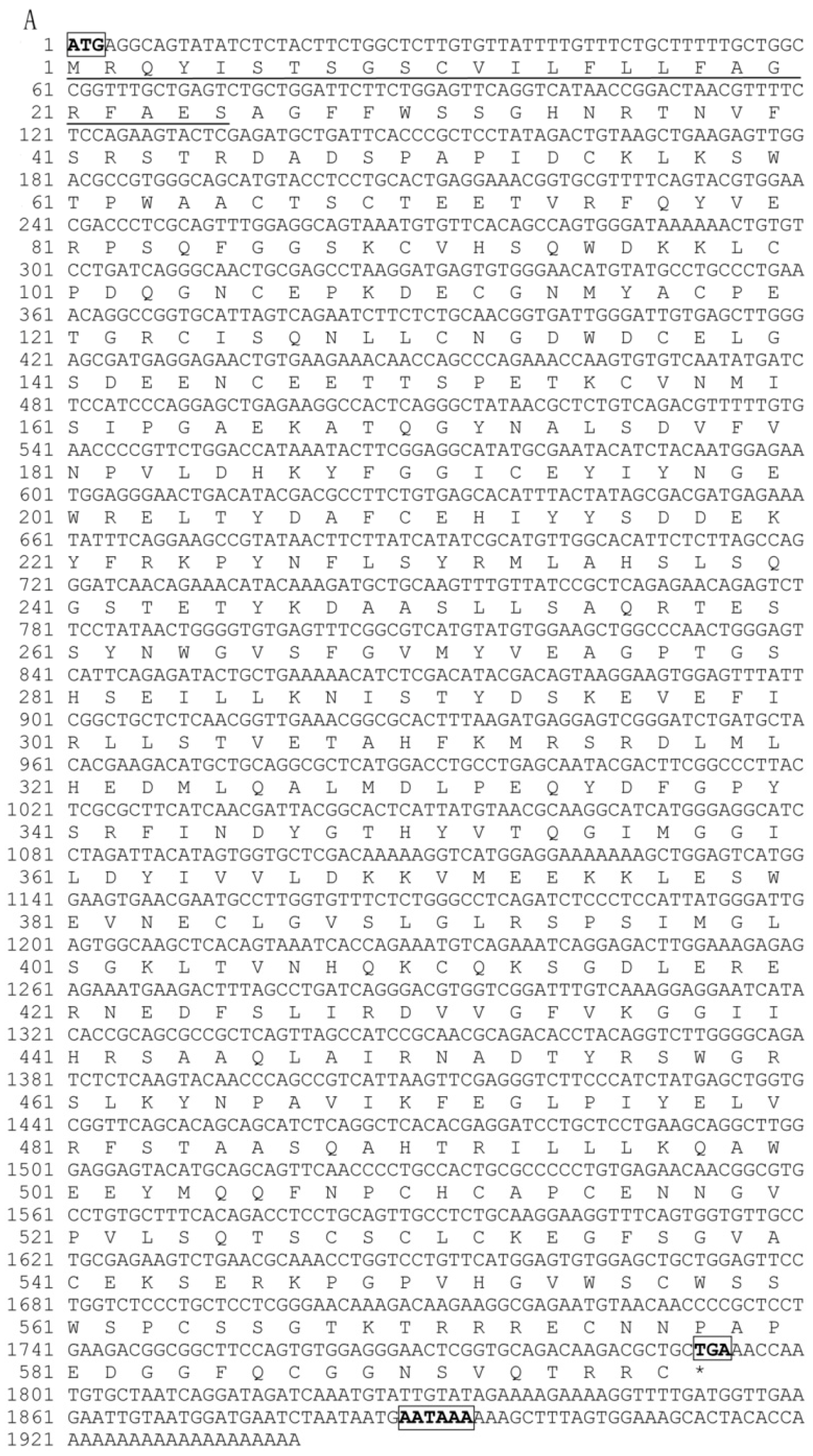
2.1. Characterization of Pf_C8α, Pf_C8β and Pf_C9 cDNA Sequences
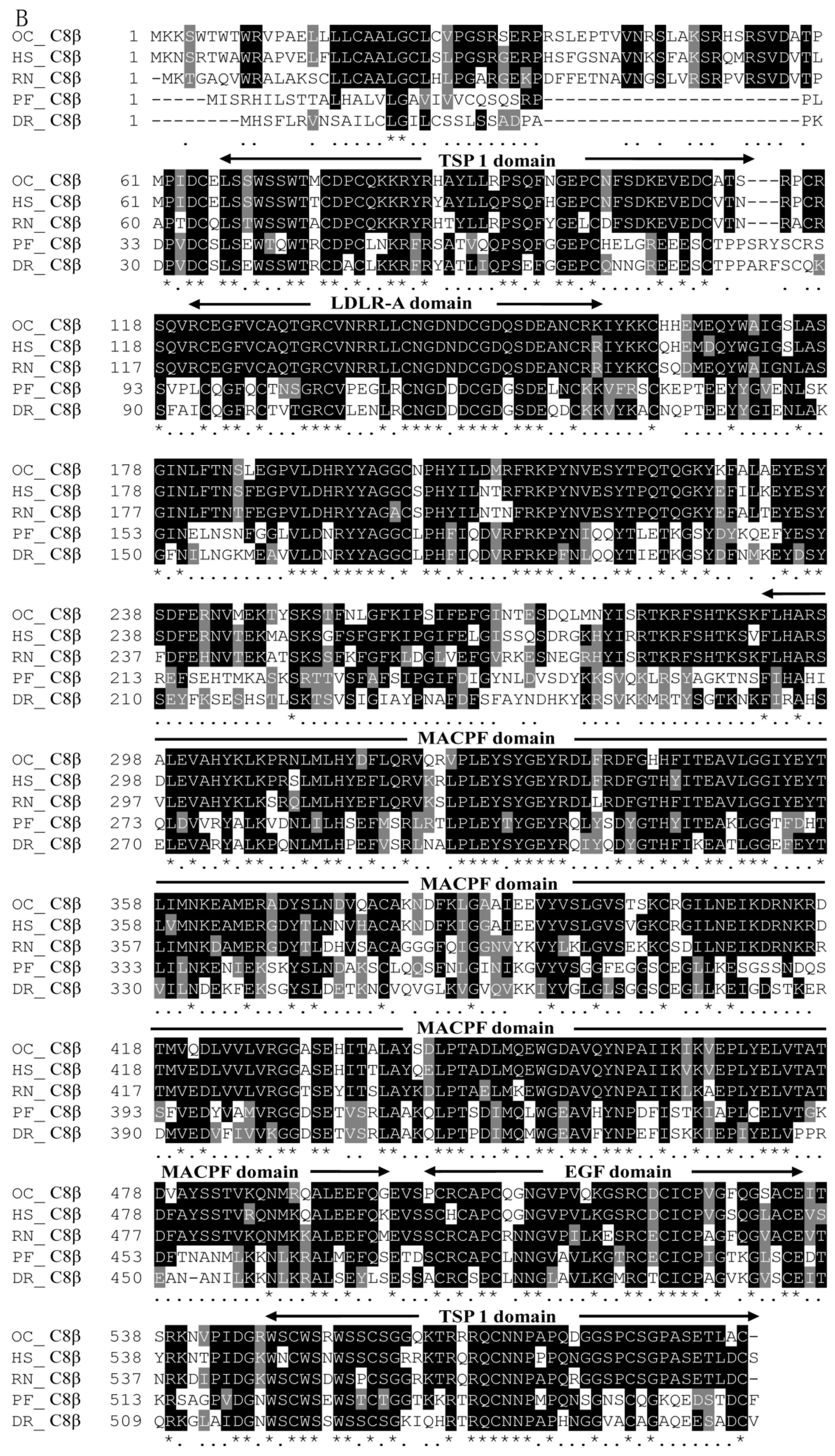
2.2. Homology Comparison and Phylogenetic Analyses of Pf_C8α, Pf_C8β and Pf_C9
2.3. Quantitative Analyses of Pf_C8α, Pf_C8β and Pf_C9 mRNA Expression Levels in Adult Tissues
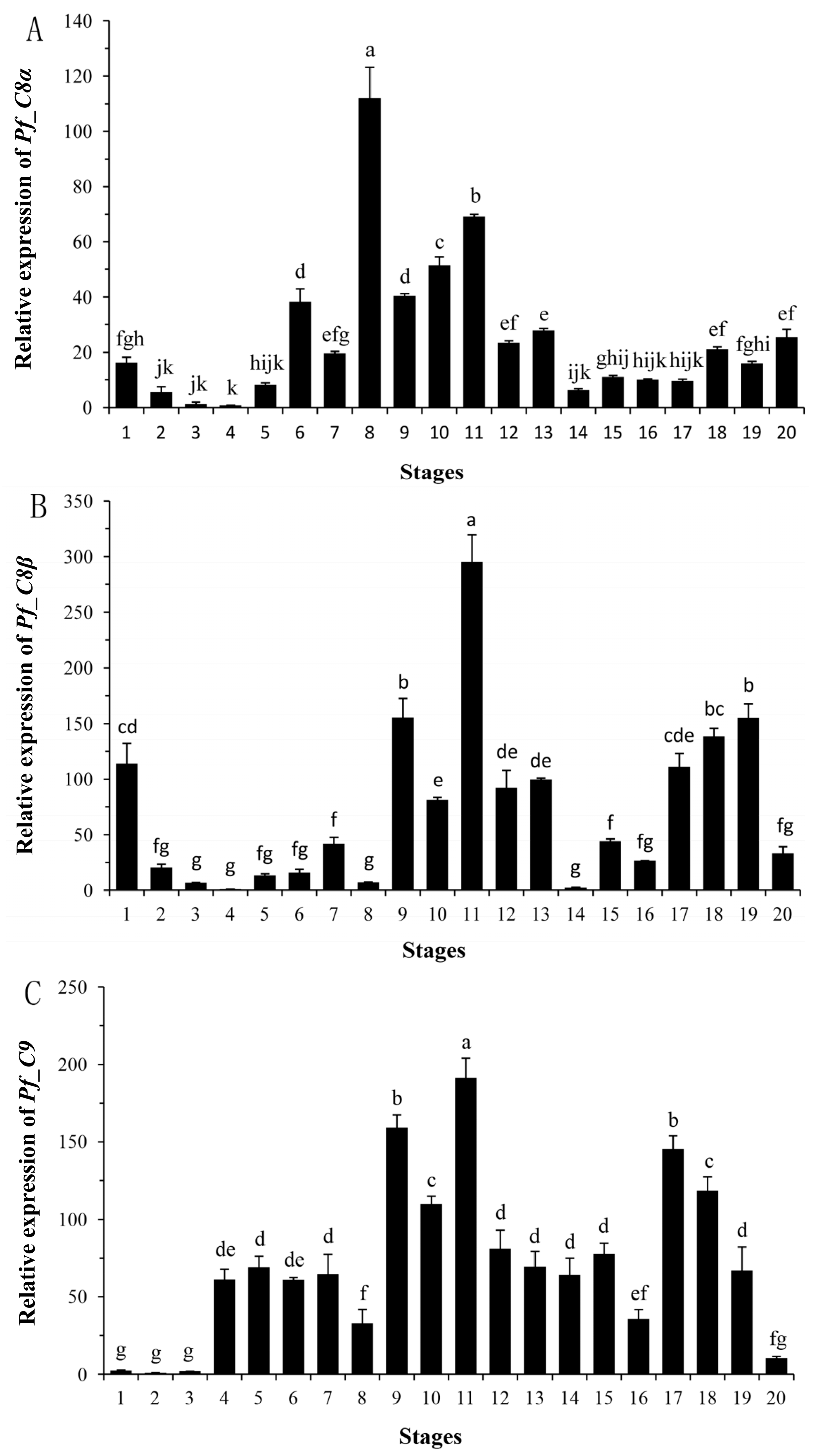
2.4. Quantitative Analyses of Pf_C8α, Pf_C8β and Pf_C9 mRNA Expression Levels during Early Developmental Stages
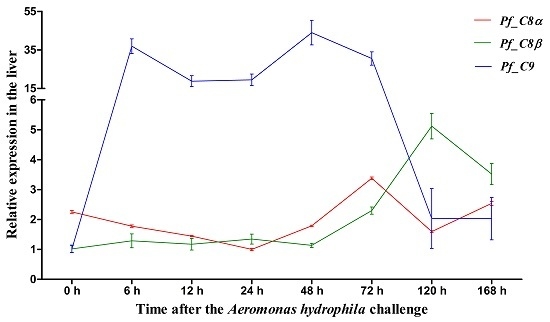
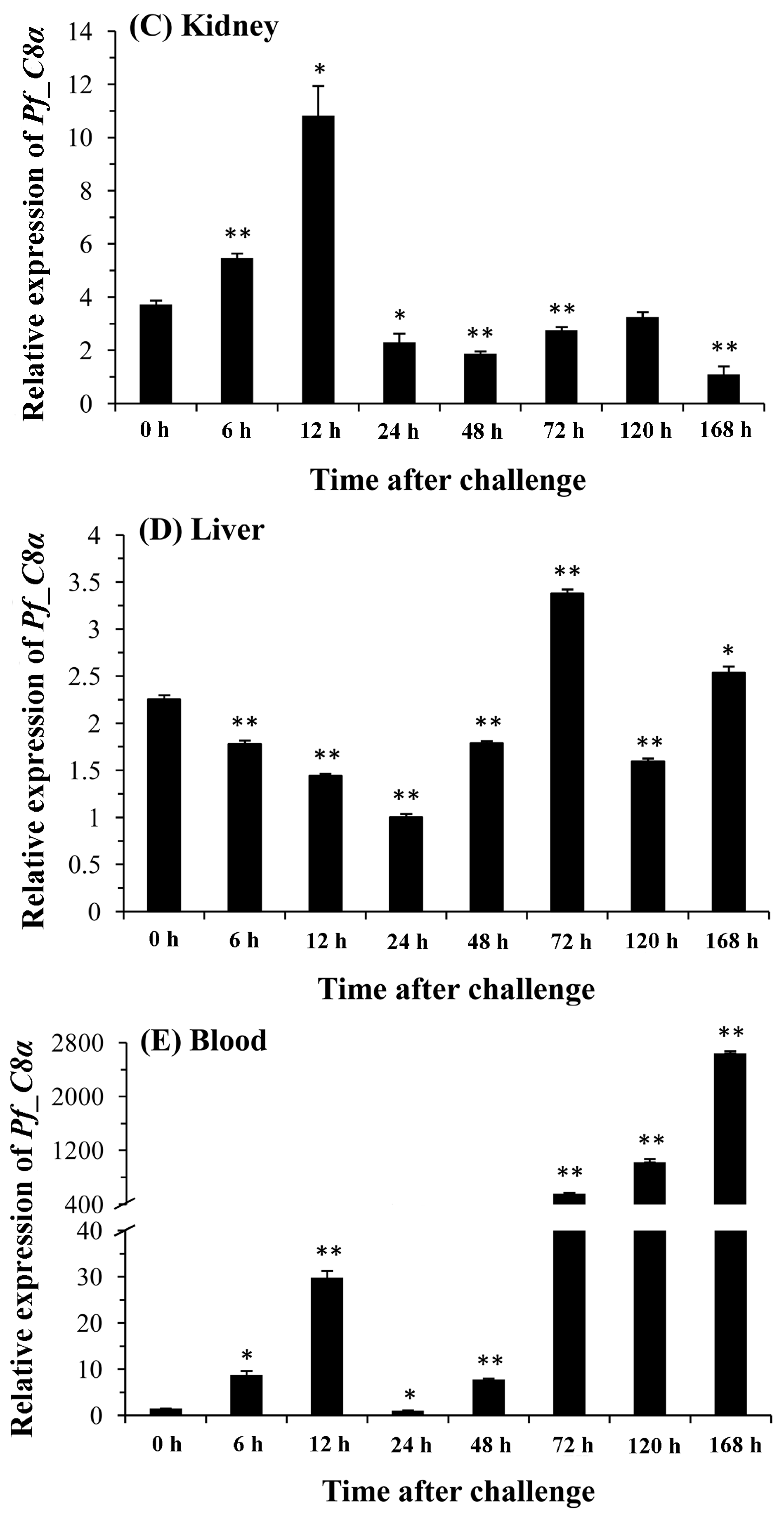
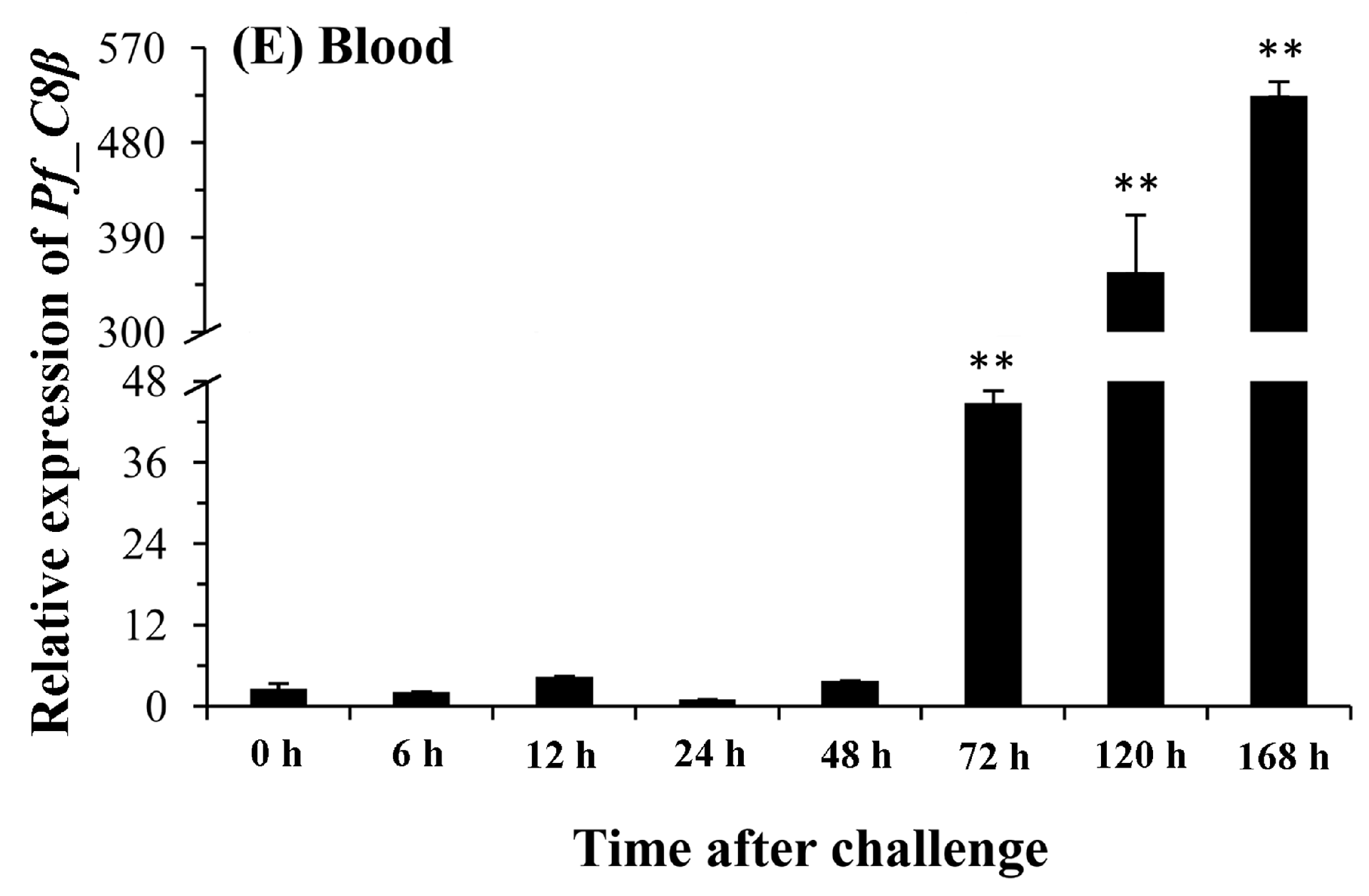
2.5. Quantitative Analysis of Pf_C8α, Pf_C8β and Pf_C9 mRNA Expression Levels in Five Tissues after Challenges of A. hydrophila
3. Discussion
4. Materials and Methods
4.1. Sampling, Bacterial Preparation and Bacterial Challenge
4.2. Total RNA Extraction and cDNA Synthesis
4.3. Cloning of cDNA Sequences for Three Genes
4.4. Sequence Analysis
4.5. qPCR and Statistical Analyses
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Carroll, M.C. The complement system in regulation of adaptive immunity. Nat. Immunol. 2004, 5, 981–986. [Google Scholar] [CrossRef] [PubMed]
- Dunkelberger, J.R.; Song, W.C. Complement and its role in innate and adaptive immune responses. Cell Res. 2010, 20, 34–50. [Google Scholar] [CrossRef] [PubMed]
- Mollnes, T.E.; Song, W.C.; Lambris, J.D. Complement in inflammatory tissue damage and disease. Trends Immunol. 2002, 23, 61–64. [Google Scholar] [CrossRef]
- Morgan, B.P.; Marchbank, K.J.; Longhi, M.P.; Harris, C.L.; Gallimore, A.M. Complement: Central to innate immunity and bridging to adaptive responses. Immunol. Lett. 2005, 97, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Wills-Karp, M. Complement activation pathways: A bridge between innate and adaptive immune responses in asthma. Proc. Am. Thorac. Soc. 2007, 4, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Rutkowski, M.J.; Sughrue, M.E.; Kane, A.J.; Ahn, B.J.; Fang, S.; Parsa, A.T. The complement cascade as a mediator of tissue growth and regeneration. Inflamm. Res. 2010, 59, 897–905. [Google Scholar] [CrossRef] [PubMed]
- Podack, E.R.; Olsen, K.J.; Lowrey, D.M.; Lichtenheld, M. Structure and function of perforin. Curr. Top. Microbiol. Immunol. 1988, 140, 11–17. [Google Scholar]
- Nonaka, M.; Kimura, A. Genomic view of the evolution of the complement system. Immunogenetics 2006, 58, 701–713. [Google Scholar] [CrossRef] [PubMed]
- Steckel, E.W.; York, R.G.; Monahan, J.B.; Sodetz, J.M. The eighth component of human complemen. J. Biol. Chem. 1980, 255, 11997–12005. [Google Scholar] [PubMed]
- Tegla, C.A.; Cudrici, C.; Patel, S.; Trippe, R.; Rus, V.; Niculescu, F.; Rus, H. Membrane attack by complement: The assembly and biology of terminal complement complexes. Immunol. Res. 2011, 51, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Parker, C.L.; Sodetz, J.M. Role of the human C8 subunits in complement-mediated bacterial killing: Evidence that C8γ is not essential. Mol. Immunol. 2002, 39, 453–458. [Google Scholar] [CrossRef]
- Hadders, M.A.; Beringer, D.X.; Gros, P. Structure of C8α-MACPF reveals mechanism of membrane attack in complement immune. Science 2007, 317, 1552–1554. [Google Scholar] [CrossRef] [PubMed]
- Monahan, J.B.; Sodetz, J.M. Role of the β subunit in interaction of the eighth component of human complement with the membrane-bound cytolytic complex. J. Biol. Chem. 1981, 256, 3258–3262. [Google Scholar] [PubMed]
- Kotnik, V.; Luznik-Bufon, T.; Schneider, P.M.; Kirschfink, M. Molecular, genetic, and functional analysis of homozygous C8β-chain deficiency in two siblings. Immunopharmacology 1997, 38, 215–221. [Google Scholar] [CrossRef]
- Luo, S.; Xie, F.; Liu, Y.; Wang, W.N. Molecular cloning, characterization and expression analysis of complement component C8 beta in the orange-spotted grouper (Epinephelus coioides) after the Vibrio alginolyticus challenge. Gene 2015, 558, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Discipio, R.G.; Berlin, C. The architectural transition of human complement component C9 to poly(C9). Mol. Immunol. 1999, 36, 575–585. [Google Scholar] [CrossRef]
- Nakao, M.; Tsujikura, M.; Ichiki, S.; Vo, T.K.; Somamoto, T. The complement system in teleost fish: Progress of post-homolog-hunting researches. Dev. Comp. Immunol. 2011, 35, 1296–1308. [Google Scholar] [CrossRef] [PubMed]
- Dishaw, L.J.; Smith, S.L.; Bigger, C.H. Characterization of a C3-like cDNA in a coral: Phylogenetic implications. Immunogenetics 2005, 57, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Thangamani, S.; Ho, B.; Ding, J.L. The ancient origin of the complement system. EMBO J. 2005, 24, 382–394. [Google Scholar] [CrossRef] [PubMed]
- Castillo, M.G.; Goodson, M.S.; McFall-Ngai, M. Identification and molecular characterization of a complement C3 molecule in a lophotrochozoan, the Hawaiian bobtail squid Euprymna scolopes. Dev. Comp. Immunol. 2009, 33, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.C.; Azumi, K.; Nonaka, M. Complement systems in invertebrates. The ancient alternative and lectin pathways. Immunopharmacology 1999, 42, 107–120. [Google Scholar] [CrossRef]
- Nonaka, M.; Azumi, K. Opsonic complement system of the solitary ascidian, Halocynthia roretzi. Dev. Comp. Immunol. 1999, 23, 421–427. [Google Scholar] [CrossRef]
- Nonaka, M.; Smith, S.L. Complement system of bony and cartilaginous fish. Fish Shellfish Immunol. 2000, 10, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Wickramaarachchi, W.D.; Wan, Q.; Lee, Y.; Lim, B.S.; de Zoysa, M.; Oh, M.J.; Jung, S.J.; Kim, H.C.; Whang, I.; Lee, J. Genomic characterization and expression analysis of complement component 9 in rock bream (Oplegnathus fasciatus). Fish Shellfish Immunol. 2012, 33, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Wickramaarachchi, W.D.; Whang, I.; Kim, E.; Lim, B.S.; Jeong, H.B.; De Zoysa, M.; Oh, M.J.; Jung, S.J.; Yeo, S.Y.; Kim, S.Y.; et al. Genomic characterization and transcriptional evidence for the involvement of complement component 7 in immune response of rock bream (Oplegnathus fasciatus). Dev. Comp. Immunol. 2013, 41, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Wickramaarachchi, W.D.; Whang, I.; Wan, Q.; Bathige, S.D.; de Zoysa, M.; Lim, B.S.; Yeo, S.Y.; Park, M.A.; Lee, J. Genomic characterization and expression analysis of complement component 8α and 8β in rock bream (Oplegnathus fasciatus). Dev. Comp. Immunol. 2013, 39, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chang, M.X.; Nie, P. Molecular cloning, promoter analysis and induced expression of the complement component C9 gene in the grass carp Ctenopharyngodon idella. Vet. Immunol. Immunopathol. 2007, 118, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Zhang, J.; Xu, X.; Fu, J.; Li, J. Expression of complement component C7 and involvement in innate immune responses to bacteria in grass carp. Fish Shellfish Immunol. 2012, 33, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Encinas, P.; Rodriguez-Milla, M.A.; Novoa, B.; Estepa, A.; Figueras, A.; Coll, J. Zebrafish fin immune responses during high mortality infections with viral haemorrhagic septicemia rhabdovirus. A proteomic and transcriptomic approach. BMC Genom. 2010, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chondrou, M.P.; Londou, A.V.; Zarkadis, I.K. Expression and phylogenetic analysis of the ninth complement component (C9) in rainbow trout. Fish Shellfish Immunol. 2006, 21, 572–576. [Google Scholar] [CrossRef] [PubMed]
- Chondrou, M.P.; Mastellos, D.; Zarkadis, I.K. cDNA cloning and phylogenetic analysis of the sixth complement component in rainbow trout. Mol. Immunol. 2006, 43, 1080–1087. [Google Scholar] [CrossRef] [PubMed]
- Zarkadis, I.K.; Duraj, S.; Chondrou, M. Molecular cloning of the seventh component of complement in rainbow trout. Dev. Comp. Immunol. 2005, 29, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Papanastasiou, A.D.; Zarkadis, I.K. Cloning and phylogenetic analysis of the α subunit of the eighth complement component (C8) in rainbow trout. Mol. Immunol. 2006, 43, 2188–2194. [Google Scholar] [CrossRef] [PubMed]
- Kazantzi, A.; Sfyroera, G.; Holland, M.C.H.; Lambris, J.D.; Zarkadis, I.K. Molecular cloning of the β subunit of complement component eight of rainbow trout. Dev. Comp. Immunol. 2003, 27, 167–174. [Google Scholar] [CrossRef]
- Shen, Y.; Zhang, J.; Xu, X.; Li, J. Molecular cloning, characterization and expression analysis of the complement component C6 gene in grass carp. Vet. Immunol. Immunopathol. 2011, 141, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Nakao, M.; Uemura, T.; Yano, T. Terminal components of carp complement constituting a membrane attack complex. Mol. Immunol. 1996, 33, 933–937. [Google Scholar] [CrossRef]
- Uemura, T.; Yano, T.; Shiraishi, H.; Nakao, M. Purification and characterization of the eighth and ninth components of carp complement. Mol. Immunol. 1996, 33, 925–932. [Google Scholar] [CrossRef]
- Zhu, L.Y.; Nie, L.; Zhu, G.; Xiang, L.X.; Shao, J.Z. Advances in research of fish immune-relevant genes: A comparative overview of innate and adaptive immunity in teleosts. Dev. Comp. Immunol. 2013, 39, 39–62. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Zhang, X.; Shi, Z.; Lin, L.; Ouyang, G.; Zhang, G.; Zheng, H.; Wei, K.; Ji, W. A Comparative transcriptome analysis between wild and albino yellow catfish (Pelteobagrus fulvidraco). PLoS ONE 2015, 10, e0131504. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.S.; Huang, J.; Shi, J.G.; Huang, Y.H.; Peng, M.Y.; Wen, H.C.; Liang, X.; Qin, J.X. Isolation and identification of ascites disease pathogen from Pelteobagrus fulvidraco Richardson and its drug sensitivity test. J. South Agric. 2012, 43, 1400–1404. [Google Scholar]
- Huang, J.; Wen, H.C.; Shi, J.G.; Huang, Y.H.; Liang, Z.S.; Long, Y.N.; Qin, J.X. Isolation and identification of pathogenic bacteria from Pelteobagrus fulvidraco ulcerative syndrome and its drug sensitive test. J. South Agric. 2012, 43, 107–112. [Google Scholar]
- Geng, Y.; Wang, K.Y.; Fan, F.L.; Chen, D.F.; Huang, J.L. Isolation, identification and biological characterization of Edwardsiella ictaluri from cultured yellow catfish Pelteobagrus fulvidraco. Oceanol. Limnol. Sin. 2010, 41, 61–67. [Google Scholar]
- Müller-Eberhard, H.J. The membrane attack complex of complement. Annu. Rev. Immunol. 1986, 4, 503–528. [Google Scholar] [CrossRef] [PubMed]
- Gasque, P. Complement: A unique innate immune sensor for danger signals. Mol. Immunol. 2004, 41, 1089–1098. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Ge, J.; Li, K.; Xu, Z.; Liang, D.; Li, J.; Li, J.; Jia, W.; Li, Y.; Dong, X.; et al. Heritable targeted inactivation of myostatin gene in yellow catfish (Pelteobagrus fulvidraco) using engineered zinc finger nucleases. PLoS ONE 2011, 6, e28897. [Google Scholar] [CrossRef] [PubMed]
- Holland, M.C.H.; Lambris, J.D. The complement system in teleosts. Fish Shellfish Immunol. 2002, 12, 399–420. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, T.; Hirono, I.; Aoki, T. Molecular analysis of complement component C8β and C9 cDNAs of Japanese flounder, Paralichthys olivaceus. Immunogenetics 1999, 50, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Witzel-Schlömp, K.; Rittner, C.; Schneider, P.M. The human complement C9 gene: Structural analysis of the 5′ gene region and genetic polymorphism studies. Eur. J. Immunogenet. 2001, 28, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, M.; Wang, C.; Ye, B.; Hua, Z. Molecular cloning of the α subunit of complement component C8 (CpC8α) of whitespotted bamboo shark (Chiloscyllium plagiosum). Fish Shellfish Immunol. 2013, 35, 1993–2000. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, S.; Su, Y.; Ye, B.; Hua, Z. Molecular characterization and expression analysis of complement component C9 gene in the whitespotted bambooshark, Chiloscyllium plagiosum. Fish Shellfish Immunol. 2013, 35, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Jeong, W.; Tian, Z. Liver: An organ with predominant innate immunity. Hepatology 2008, 47, 729–736. [Google Scholar] [CrossRef] [PubMed]
- Magnadóttir, B. Innate immunity of fish (overview). Fish Shellfish Immunol. 2006, 20, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Li, C.M. Cloning and Expression Analysis of Terminal Complement Components of Cyprinus carpio haematopteru. Master Thesis, Henan Normal University, Xinxiang, China, May 2012. [Google Scholar]
- Mulero, I.; García-Ayala, A.; Meseguer, J.; Mulero, V. Maternal transfer of immunity and ontogeny of autologous immunocompetence of fish: A minireview. Aquaculture 2007, 268, 244–250. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, S.; Wang, G.; An, Y. Complement activity in the egg cytosol of zebrafish Danio rerio: Evidence for the defense role of maternal complement components. PLoS ONE 2008, 1, e1463. [Google Scholar] [CrossRef] [PubMed]
- Lechniak, D.; Pers-Kamczyc, E.; Pawlak, P. Timing of the first zygotic cleavage as a marker of developmental potential of mammalian embryos. Reprod. Biol. 2008, 8, 23–42. [Google Scholar] [CrossRef]
- Liang, T.; Ji, W.; Zhang, G.R.; Wei, K.J.; Feng, K.; Wang, W.M.; Zou, G.W. Molecular cloning and expression analysis of liver-expressed antimicrobial peptide 1 (LEAP-1) and LEAP-2 genes in the blunt snout bream (Megalobrama amblycephala). Fish Shellfish Immunol. 2013, 35, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real time quantitative PCR and the 2−ΔΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]










| Protein | Species | GenBank Accession no. |
|---|---|---|
| C8α | Esox lucius | XP_010885181.1 |
| Oncorhynchus mykiss | NP_001118096.1 | |
| Oplegnathus fasciatus | AFZ93888.1 | |
| Siniperca chuatsi | AKA66305.1 | |
| Clupea harengus | XP_012681433.1 | |
| Pelteobagrus fulvidraco | KT588317 | |
| Danio rerio | AAH78409.1 | |
| Xenopus (Silurana) tropicalis | AAH74554.1 | |
| Gallus gallus | XP_426667.2 | |
| Mus musculus | AAH27748.1 | |
| Bos taurus | AAI12636.1 | |
| Homo sapiens | AAI32914.1 | |
| Oryctolagus cuniculus | NP_001075724.1 | |
| C8β | Homo sapiens | NP_000057.2 |
| Macaca mulatta | XP_001114456.1 | |
| Oryctolagus cuniculus | P98137.1 | |
| Rattus norvegicus | NP_001178688.1 | |
| Pelteobagrus fulvidraco | KT588318 | |
| Danio rerio | NP_001243652.1 | |
| Esox lucius | XP_010885201.1 | |
| Oncorhynchus mykiss | AAL16647.1 | |
| Neolamprologus brichardi | XP_006786991.1 | |
| Paralichthys olivaceus | BAA86877.1 | |
| C9 | Oryctolagus cuniculus | NP_001075815.1 |
| Homo sapiens | AAH20721.1 | |
| Sus scrofa | NP_001090917.1 | |
| Rattus norvegicus | AAB38023.1 | |
| Xenopus (Silurana) tropicalis | AAI59018.1 | |
| Danio rerio | XP_005171925.1 | |
| Ctenopharyngodon idella | ABN49522.1 | |
| Pelteobagrus fulvidraco | KT454382 | |
| Plecoglossus altivelis | CBX31962.1 | |
| Oncorhynchus mykiss | NP_001117898.1 | |
| Xiphophorus hellerii | AEJ08068.1 | |
| Oplegnathus fasciatus | AFU81223.1 | |
| Paralichthys olivaceus | BAA86878.1 | |
| Takifugu rubripes | AAC60288.1 |
| Name of Primer | Sequence of Primes (5′→3′) | Ta (°C) | Application |
|---|---|---|---|
| C8α-F | ATGAGGCAGTATATCTCTACTTCTGG | 60 | cDNA cloning of C8α gene |
| C8α-R | TCAGCAGCGTCTTGTCTGCA | ||
| C8α-3′RACE outer | CTGTCAGATGGCGGGTGGCA | 58 | 3′RACE (1st round PCR) |
| C8α-3′RACE inner | CTTCAGTTCGCCTGCTCCCG | 58 | 3′RACE (2nd round PCR) |
| C8β-F1 | CGCCGTCACGCTACTCAT | 60 | cDNA Cloning of C8β gene |
| C8β-R1 | GTGGTCAAAGGTTCCCCC | ||
| C8β-F2 | CTTTCGTTTCGTGCCT | 50 | cDNA cloning of C8β gene |
| C8β-R2 | GTGTATTGCTGGATGTTGTA | ||
| C8β-3′RACE outer | CCTGCGAACTCTCCCACT | 55 | 3′RACE (1st round PCR) |
| C8β-3′RACE inner | GCTTGGGGGAACCTTTGAC | 55 | 3′RACE (2nd round PCR) |
| C9-F | ATGAGGATGTTGATGTCAAT | 56 | cDNA cloning of C9 gene |
| C9-R | TTAACAATATTCGTCGCTGA | ||
| C9-3′RACE outer | CACCCCTACTGAGTCATCGG | 58 | 3′RACE (1st round PCR) |
| C9-3′RACE inner | ATTACAGACCACAGCACCAC | 54 | 3′RACE (2nd round PCR) |
| Oligo(dT)17 | GACTCGAGTCGACATCGA(T)17 | Universal primer for 3′RACE | |
| linker adapter | GACTCGAGTCGACATCG | Universal primer for 3′RACE |
| Name of Primer | Sequence of Primes (5′→3′) | Ta (°C) | Application |
|---|---|---|---|
| C8α-DLF | TGCCTGAGCAATACGACT | 58 | qPCR of C8α mRNA |
| C8α-DLR | CGAGCACCACTATGTAATCTA | ||
| C8β-DLF | CAAGGAGCCAACAGAAGA | 55 | qPCR of C8β mRNA |
| C8β-DLR | TGAAGGCAAAGGAGACAG | ||
| C9-DLF | TTCATTGCTGAGCGAACT | 62 | qPCR of C9 mRNA |
| C9-DLR | ATTTGTGGTGCTGTGGTC | ||
| β-actin-F | TCCCTGTATGCCTCTGGTGGT | 58 | qPCR of β-actin mRNA |
| β-actin-R | AAGCTGTAGCCTCTCTCGGTC |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, H.; Ji, W.; Zhang, G.-R.; Zhang, X.-T.; Shi, Z.-C.; Wei, K.-J.; Yang, R.-B.; Gardner, J.P.A. Molecular Characterization and Expression Analyses of the Complement Component C8α, C8β and C9 Genes in Yellow Catfish (Pelteobagrus fulvidraco) after the Aeromonas hydrophila Challenge. Int. J. Mol. Sci. 2016, 17, 345. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17030345
Zheng H, Ji W, Zhang G-R, Zhang X-T, Shi Z-C, Wei K-J, Yang R-B, Gardner JPA. Molecular Characterization and Expression Analyses of the Complement Component C8α, C8β and C9 Genes in Yellow Catfish (Pelteobagrus fulvidraco) after the Aeromonas hydrophila Challenge. International Journal of Molecular Sciences. 2016; 17(3):345. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17030345
Chicago/Turabian StyleZheng, Huan, Wei Ji, Gui-Rong Zhang, Xiao-Ting Zhang, Ze-Chao Shi, Kai-Jian Wei, Rui-Bin Yang, and Jonathan P. A. Gardner. 2016. "Molecular Characterization and Expression Analyses of the Complement Component C8α, C8β and C9 Genes in Yellow Catfish (Pelteobagrus fulvidraco) after the Aeromonas hydrophila Challenge" International Journal of Molecular Sciences 17, no. 3: 345. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17030345