Molecular Cloning and Expression Analysis of IgD in Nile Tilapia (Oreochromis niloticus) in Response to Streptococcus agalactiae Stimulus
Abstract
:1. Introduction
2. Results
2.1. Characterization of On-mIgD
2.2. Homology Search
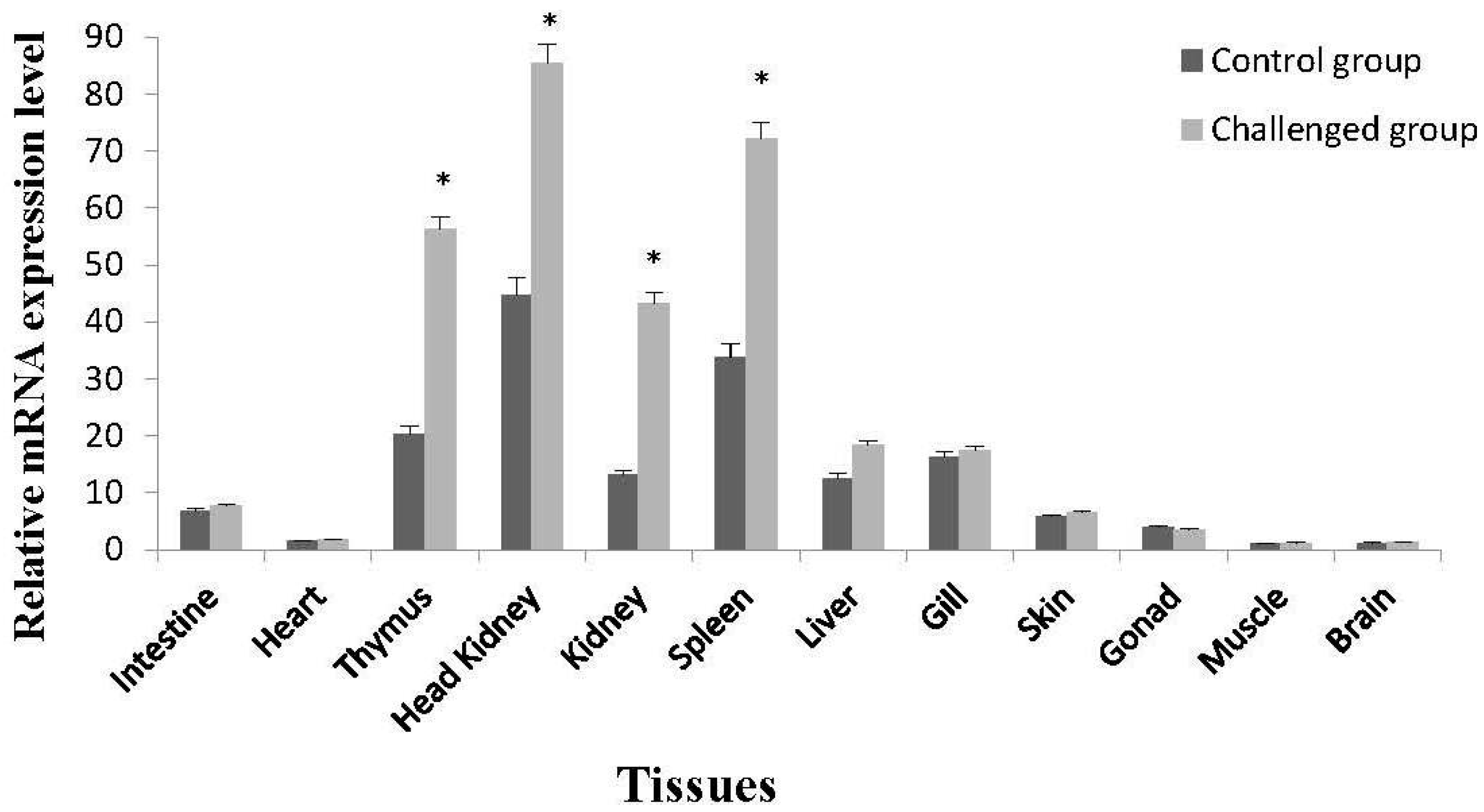
2.3. Tissue-Specific Expression of On-mIgD mRNA
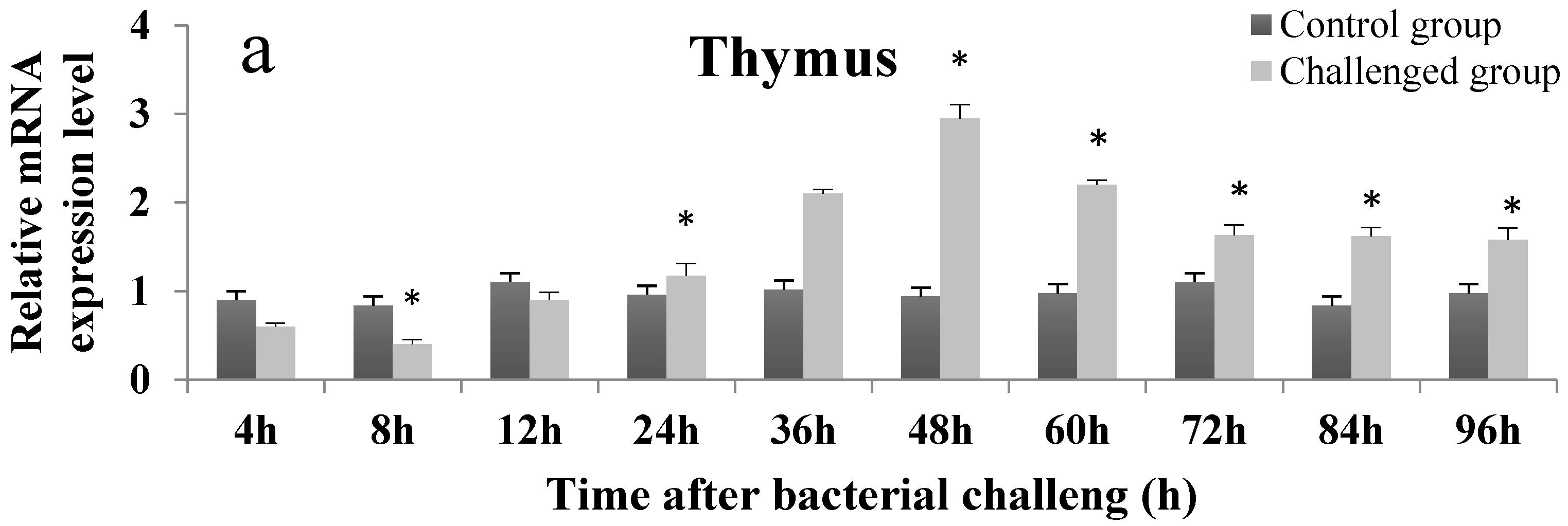
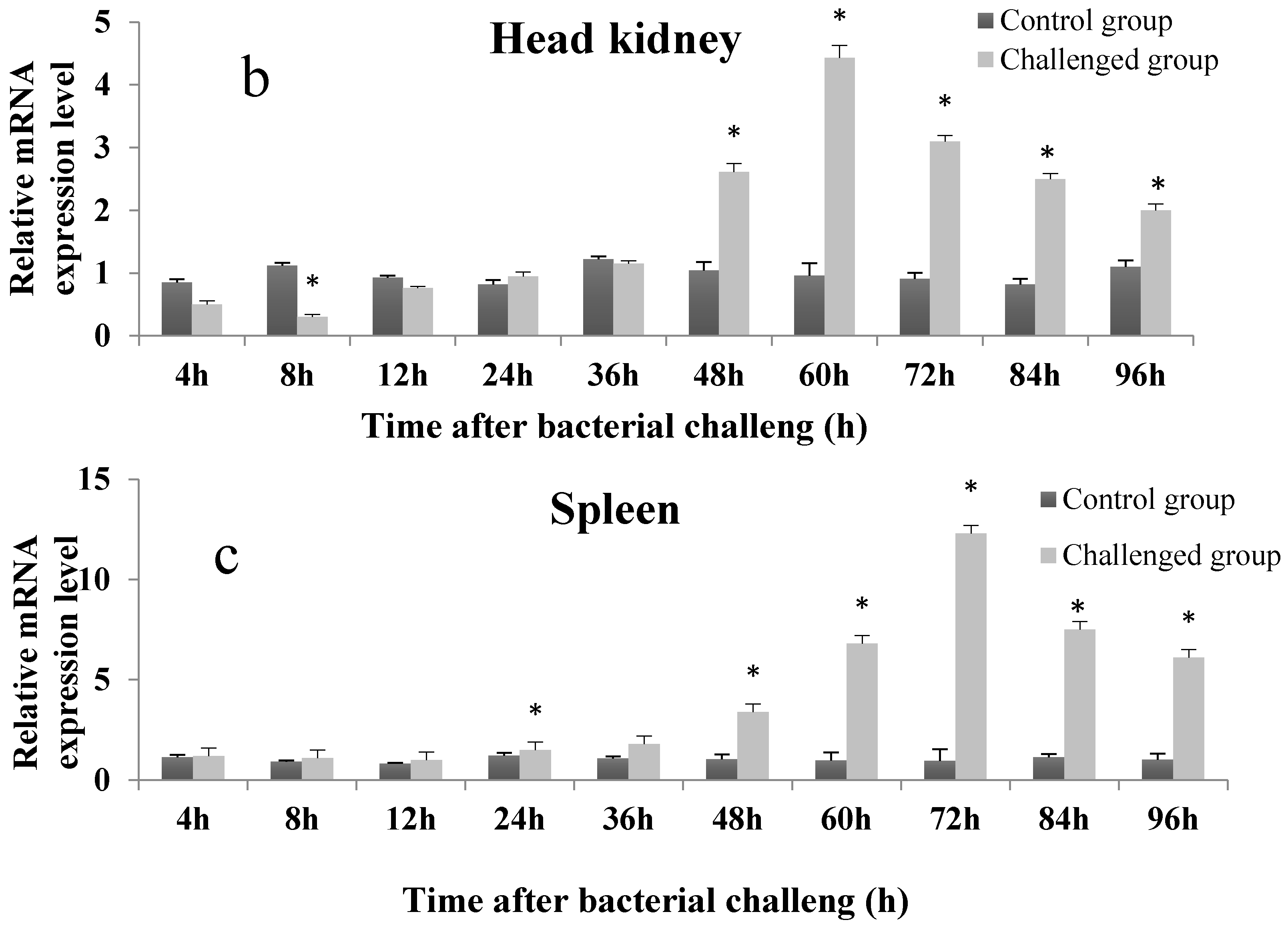
2.4. Temporal Expression Profile of On-mIgD mRNA
3. Discussion
4. Materials and Methods
4.1. Animals and Preparation
4.2. Amplification of IgD cDNA
4.3. 3′ RACE
4.4. 5′ RACE
4.5. Bioinformations Analysis
4.6. S. agalactiae Challenge of the Fish
4.7. Tissue-Specific Expression of On-mIgD mRNA
4.8. The Temporal Expression of On-mIgD mRNA in Head Kidney, Thymus, and Spleen after Bacterial Challenge
4.9. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wilson, M.; Bengtén, E.; Miller, N.W.; Clem, L.W.; Du Pasquier, L.; Warr, G.W. A novel chimeric ig heavy chain from a teleost fish shares similarities to IgD. Proc. Natl. Acad. Sci. USA 1997, 94, 4593–4597. [Google Scholar] [CrossRef] [PubMed]
- Hordvik, I.; Thevarajan, J.; Samdal, I.; Bastani, N.; Krossoy, B. Molecular cloning and phylogenetic analysis of the atlantic salmon immunoglobulin d gene. Scand. J. Immunol. 1999, 50, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Stenvik, J.; Jørgensen, T.Ø. Immunoglobulin D (IgD) of atlantic cod has a unique structure. Immunogenetics 2000, 51, 452–461. [Google Scholar] [CrossRef] [PubMed]
- Hirono, I.; Nam, B.-H.; Enomoto, J.; Uchino, K.; Aoki, T. Cloning and characterisation of a cDNA encoding Japanese flounder paralichthys olivaceus IgD. Fish Shellfish Immunol. 2003, 15, 63–70. [Google Scholar] [CrossRef]
- Savan, R.; Aman, A.; Nakao, M.; Watanuki, H.; Sakai, M. Discovery of a novel immunoglobulin heavy chain gene chimera from common carp (Cyprinus carpio L.). Immunogenetics 2005, 57, 458–463. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, A.M.; Moustafa, F.M.; Romanowski, K.E.; Steiner, L.A. Zebrafish immunoglobulin IgD: Unusual exon usage and quantitative expression profiles with IgM and IgZ/T heavy chain isotypes. Mol. Immunol. 2011, 48, 2220–2223. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Gomez, F.; Greene, W.; Rego, K.; Hansen, J.D.; Costa, G.; Kataria, P.; Bromage, E.S. Discovery and characterization of secretory IgD in rainbow trout: Secretory IgD is produced through a novel splicing mechanism. J. Immunol. 2012, 188, 1341–1349. [Google Scholar] [CrossRef] [PubMed]
- Saha, N.R.; Suetake, H.; Kikuchi, K.; Suzuki, Y. Fugu immunoglobulin d: A highly unusual gene with unprecedented duplications in its constant region. Immunogenetics 2004, 56, 438–447. [Google Scholar] [CrossRef] [PubMed]
- White, M.B.; Shen, A.L.; Word, C.J.; Tucker, P.W.; Blattner, F.R. Human immunoglobulin D: Genomic sequence of the delta heavy chain. Science 1985, 228, 733–737. [Google Scholar] [CrossRef] [PubMed]
- Tucker, P.W.; Liu, C.-P.; Mushinski, J.F.; Blattner, F.R. Mouse immunoglobulin D: Messenger RNA and genomic DNA sequences. Science 1980, 209, 1353–1360. [Google Scholar] [CrossRef] [PubMed]
- Hordvik, I. Identification of a novel immunoglobulin δ transcript and comparative analysis of the genes encoding IgD in atlantic salmon and atlantic halibut. Mol. Immunol. 2002, 39, 85–91. [Google Scholar] [CrossRef]
- Bengtén, E.; Quiniou, S.M.-A.; Stuge, T.B.; Katagiri, T.; Miller, N.W.; Clem, L.W.; Warr, G.W.; Wilson, M. The IgH locus of the channel catfish, ictalurus punctatus, contains multiple constant region gene sequences: Different genes encode heavy chains of membrane and secreted IgD. J. Immunol. 2002, 169, 2488–2497. [Google Scholar] [CrossRef] [PubMed]
- Robinette, D.; Wada, S.; Arroll, T.; Levy, M.; Miller, W.; Noga, E. Antimicrobial activity in the skin of the channel catfish ictalurus punctatus: Characterization of broad-spectrum histone-like antimicrobial proteins. Cell. Mol. Life Sci. CMLS 1998, 54, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Fırat, Ö.; Cogun, H.Y.; Yüzereroğlu, T.A.; Gök, G.; Fırat, Ö.; Kargin, F.; Kötemen, Y. A comparative study on the effects of a pesticide (cypermethrin) and two metals (copper, lead) to serum biochemistry of Nile tilapia, oreochromis niloticus. Fish Physiol. Biochem. 2011, 37, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Lukkana, M.; Jantrakajorn, S.; Wongtavatchai, J. Antimicrobial susceptibility and enrofloxacin resistance of streptococcal bacteria from farmed nile tilapia, oreochromis niloticus (linnaeus 1758) in Thailand. Aquac. Res. 2015. [Google Scholar] [CrossRef]
- Chen, M.; Li, L.-P.; Wang, R.; Liang, W.-W.; Huang, Y.; Li, J.; Lei, A.-Y.; Huang, W.-Y.; Gan, X. PCR detection and PFGE genotype analyses of streptococcal clinical isolates from tilapia in China. Vet. Microbiol. 2012, 159, 526–530. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Jian, J.; Lu, Y.; Cai, S.; Huang, Y.; Tang, J.; Wu, Z. Complete genome sequence of Streptococcus agalactiae ZQ0910, a pathogen causing meningoencephalitis in the gift strain of nile tilapia (Oreochromis niloticus). J. Bacteriol. 2012, 194, 5132–5133. [Google Scholar] [CrossRef] [PubMed]
- Stenvik, J.; Schrøder, M.; Olsen, K.; Zapata, A.; Jørgensen, T.Ø. Expression of immunoglobulin heavy chain transcripts (VH-families, IgM, and IgD) in head kidney and spleen of the atlantic cod (Gadus morhua L.). Dev. Comparat. Immunol. 2001, 25, 291–302. [Google Scholar] [CrossRef]
- Tian, J.; Sun, B.; Luo, Y.; Zhang, Y.; Nie, P. Distribution of IgM, IgD and IgZ in mandarin fish, Siniperca chuatsi lymphoid tissues and their transcriptional changes after flavobacterium columnare stimulation. Aquaculture 2009, 288, 14–21. [Google Scholar] [CrossRef]
- Romano, N.; Ceccariglia, S.; Mastrolia, L.; Mazzini, M. Cytology of lymphomyeloid head kidney of antarctic fishes Trematomus bernacchii (nototheniidae) and Chionodraco hamatus (channicthyidae). Tissue Cell 2002, 34, 63–72. [Google Scholar] [CrossRef]
- Mebius, R.E.; Kraal, G. Structure and function of the spleen. Nat. Rev. Immunol. 2005, 5, 606–616. [Google Scholar] [CrossRef] [PubMed]
- Cesta, M.F. Normal structure, function, and histology of the spleen. Toxicol. Pathol. 2006, 34, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Gan, Z.; Wang, B.; Lu, Y.; Cai, S.; Cai, J.; Jian, J.; Wu, Z. Molecular characterization and expression of CD2BP2 in nile tilapia (Oreochromis niloticus) in response to streptococcus agalactiae stimulus. Gene 2014, 548, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Doran, K.S.; Engelson, E.J.; Khosravi, A.; Maisey, H.C.; Fedtke, I.; Equils, O.; Michelsen, K.S.; Arditi, M.; Peschel, A.; Nizet, V. Blood-brain barrier invasion by group B Streptococcus depends upon proper cell-surface anchoring of lipoteichoic acid. J. Clin. Investig. 2005, 115, 2499. [Google Scholar] [CrossRef] [PubMed]
- Eldar, A.; Bejerano, Y.; Livoff, A.; Horovitcz, A.; Bercovier, H. Experimental streptococcal meningo-encephalitis in cultured fish. Vet. Microbiol. 1995, 43, 33–40. [Google Scholar] [CrossRef]
- Hunolstein, V. Group B streptococci persist inside macrophages. Immunology 1998, 93, 86–95. [Google Scholar]
- Nizet, V.; Kim, K.; Stins, M.; Jonas, M.; Chi, E.Y.; Nguyen, D.; Rubens, C.E. Invasion of brain microvascular endothelial cells by group B streptococci. Infect. Immunity 1997, 65, 5074–5081. [Google Scholar]
- Valenti-Weigand, P.; Benkel, P.; Rohde, M.; Chhatwal, G.S. Entry and intracellular survival of group B streptococci in J774 macrophages. Infect. Immun. 1996, 64, 2467–2473. [Google Scholar]
- Tha-In, T.; Bayry, J.; Metselaar, H.J.; Kaveri, S.V.; Kwekkeboom, J. Modulation of the cellular immune system by intravenous immunoglobulin. Trends Immunol. 2008, 29, 608–615. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.L.; Jian, J.C.; Lu, Y.S.; Wang, B.; Wu, Z.H. A tandem-repeat galectin involved in innate immune response of the pearl oyster Pinctada fucata. Marin Genom. 2011, 4, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]






| Primers | Sequence (5′–3′) |
|---|---|
| Oligo dT30-anchor | AAGCAGTGGTATCAACGCAGAGTACT(30)VN |
| DF1 | GGAGTCAGTCAARTCCRAGT |
| DR1 | CTGCTYAGRCTGAASGTTTT |
| DSP1 | CGTTGTTTTGATTCAGTGGTTGT |
| DSP2 | AGATGAGCCTTACTGCTGAGATT |
| DSP3 | AAGCAGTGGTATCAACGCAGAGT |
| Oligo dT-anchor | GACCACGCGTATCGATGTCGACT(16) V |
| Anchor primer | GACCACGCGTATCGATGTCGAC |
| QrD-F | AACACCACCCTGTCCCTGAAT |
| QrD-R | GGGTGAAAACCACATTCCAGC |
| β-action-F | AACAACCACACACCACACATTTC |
| β-action-R | TGTCTCCTTCATCGTTCCAGTTT |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.; Wang, P.; Wu, Z.-H.; Lu, Y.-S.; Wang, Z.-L.; Jian, J.-C. Molecular Cloning and Expression Analysis of IgD in Nile Tilapia (Oreochromis niloticus) in Response to Streptococcus agalactiae Stimulus. Int. J. Mol. Sci. 2016, 17, 348. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17030348
Wang B, Wang P, Wu Z-H, Lu Y-S, Wang Z-L, Jian J-C. Molecular Cloning and Expression Analysis of IgD in Nile Tilapia (Oreochromis niloticus) in Response to Streptococcus agalactiae Stimulus. International Journal of Molecular Sciences. 2016; 17(3):348. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17030348
Chicago/Turabian StyleWang, Bei, Pei Wang, Zao-He Wu, Yi-Shan Lu, Zhong-Liang Wang, and Ji-Chang Jian. 2016. "Molecular Cloning and Expression Analysis of IgD in Nile Tilapia (Oreochromis niloticus) in Response to Streptococcus agalactiae Stimulus" International Journal of Molecular Sciences 17, no. 3: 348. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17030348