LARP6 Meets Collagen mRNA: Specific Regulation of Type I Collagen Expression
Abstract
:1. Fibrotic Diseases
2. Type I Collagen
3. Posttranscriptional Regulation of Expression of Type I Collagen
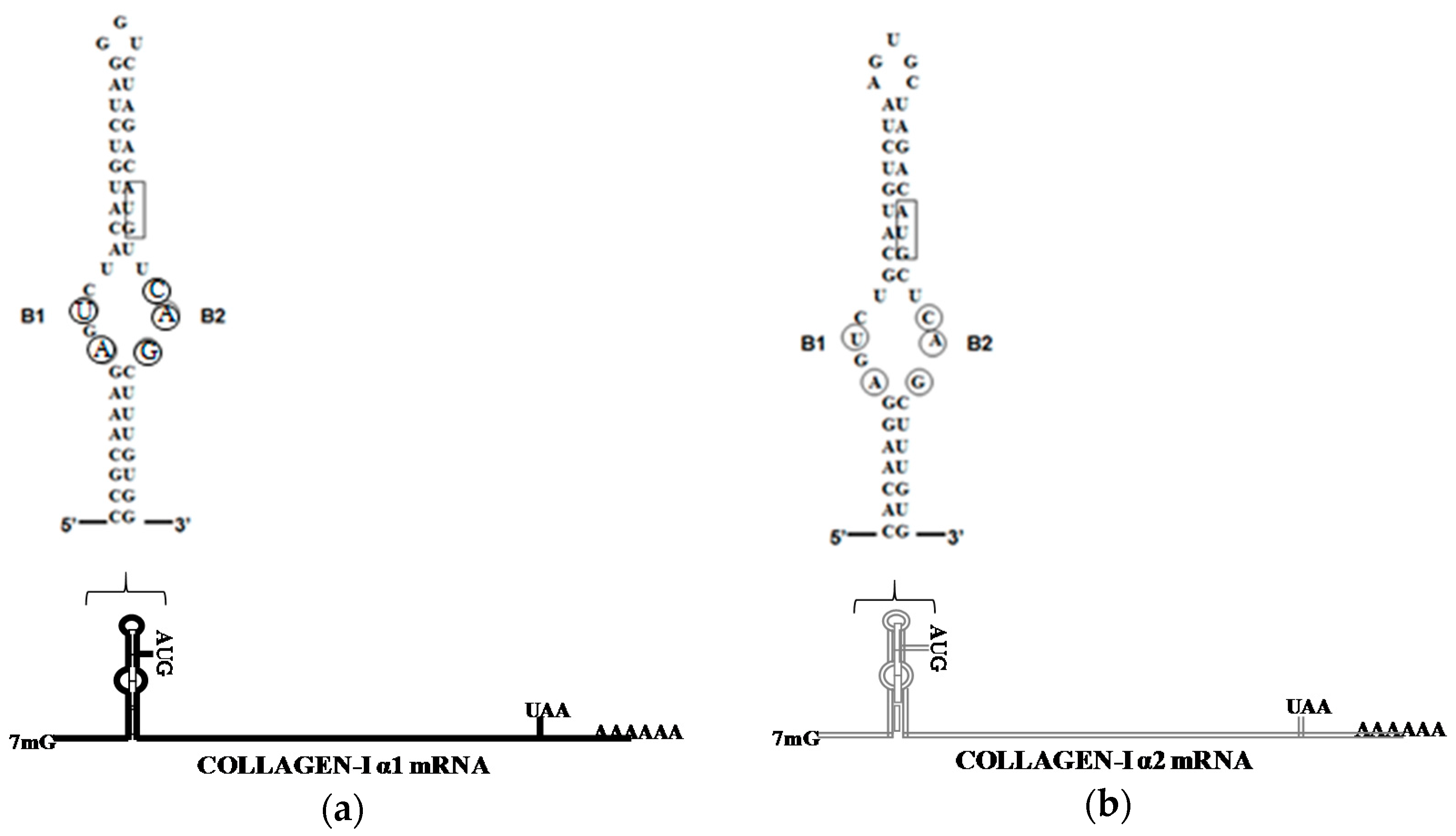
4. LARP6 Is the Specific RNA Binding Protein of Collagen mRNAs
5. Role of LARP6 Binding in Fibrosis Development
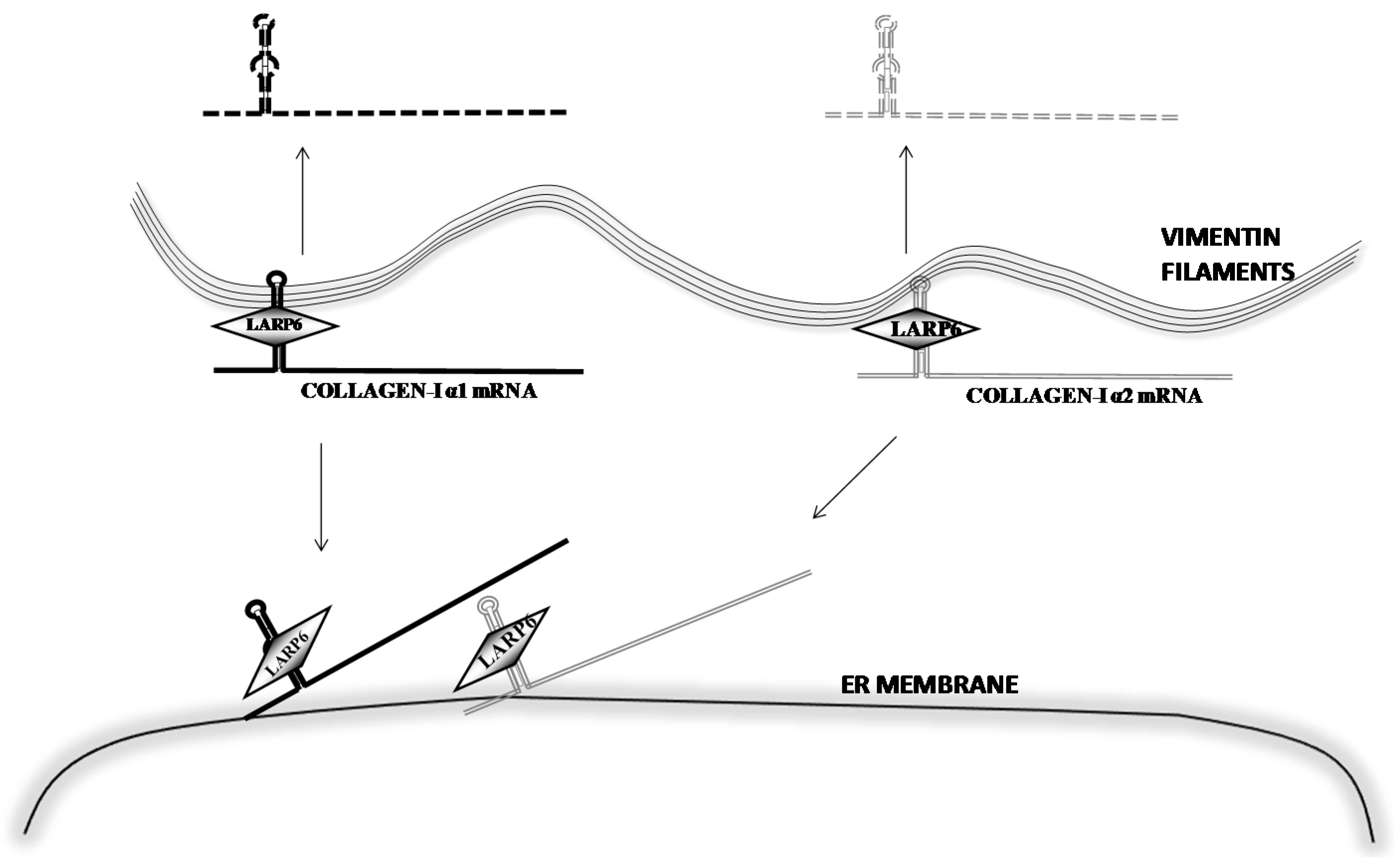
6. Stabilization of Collagen mRNAs by Interaction of LARP6 and Vimentin Filaments
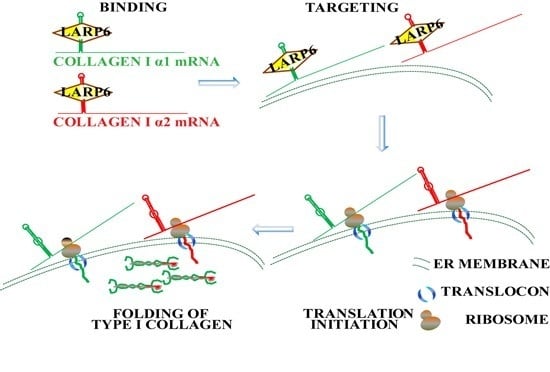
7. Unique Aspects of Translation of Collagen mRNAs
8. Coordinated Translation of Collagen mRNAs Is Regulated by Interaction of LARP6 and Serine Threonine Kinase Receptor Associated Protein (STRAP)
9. RNA Helicase A Increases Translational Competitiveness of Collagen mRNA
10. Interaction of LARP6 and FKBP3 Increases the Half-Life of LARP6
11. Phosphorylation of LARP6 Regulates Its Activity in Fibrosis
12. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| LARP6 | La ribonucleoproteins domain family, member 6 |
| ER | endoplasmic reticulum |
| HSC | hepatic stellate cell |
| TGF-β | transforming growth factor β |
| RRM | RNA recognition motif |
| nt | nucleotides |
| LOX | 15-lipoxygenase |
| ORF | open reading frame |
| UNRIP | upstream of N-ras interacting protein |
| STRAP | serine threonine kinase receptor associated protein |
| RHA | RNA helicase A |
| PCE | posttranscriptional control element |
| FKBP25/FKBP3 | 25 kD FK506 binding protein |
| PPIase | cis-trans prolyl isomerase |
| CTER | C-terminal of domain |
References
- Zeisberg, M.; Neilson, E.G. Mechanisms of tubulointerstitial fibrosis. J. Am. Soc. Nephrol. 2010, 21, 1819–1834. [Google Scholar] [CrossRef] [PubMed]
- Shahbaz, A.U.; Sun, Y.; Bhattacharya, S.K.; Ahokas, R.A.; Gerling, I.C.; McGee, J.E.; Weber, K.T. Fibrosis in hypertensive heart disease: Molecular pathways and cardioprotective strategies. J. Hypertens. 2010, 28, S25–S32. [Google Scholar] [CrossRef] [PubMed]
- Rombouts, K.; Marra, F. Molecular mechanisms of hepatic fibrosis in non-alcoholic steatohepatitis. Dig. Dis. 2010, 28, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Pinzani, M.; Macias-Barragan, J. Update on the pathophysiology of liver fibrosis. Expert Rev. Gastroenterol. Hepatol. 2010, 4, 459–472. [Google Scholar] [CrossRef] [PubMed]
- Trojanowska, M.; LeRoy, E.C.; Eckes, B.; Krieg, T. Pathogenesis of fibrosis: Type 1 collagen and the skin. J. Mol. Med. 1998, 76, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Longo, D.L.; Rockey, D.C.; Bell, P.D.; Hill, J.A. Fibrosis—A common pathway to organ injury and failure. N. Engl. J. Med. 2015, 372, 1138–1149. [Google Scholar] [CrossRef] [PubMed]
- Ricard-Blum, S.; Ruggiero, F. The collagen superfamily: From the extracellular matrix to the cell membrane. Pathol. Biol. (Paris). 2005, 53, 430–442. [Google Scholar] [CrossRef] [PubMed]
- Van der Rest, M.; Garrone, R. Collagen family of proteins. FASEB J. 1991, 5, 2814–2823. [Google Scholar] [PubMed]
- Gelse, K.; Pöschl, E.; Aigner, T. Collagens—Structure, function, and biosynthesis. Adv. Drug Deliv. Rev. 2003, 55, 1531–1546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, K.; Rennie, M.J. New approaches and recent results concerning human-tissue collagen synthesis. Curr. Opin. Clin. Nutr. Metab. Care 2007, 10, 582–590. [Google Scholar] [CrossRef] [PubMed]
- Rucklidge, G.J.; Milne, G.; McGaw, B.A.; Milne, E.; Robins, S.P. Turnover rates of different collagen types measured by isotope ratio mass spectrometry. BBA Gen. Subj. 1992, 1156, 57–61. [Google Scholar] [CrossRef]
- Nissen, R.; Cardinale, G.J.; Udenfriend, S. Increased turnover of arterial collagen in hypertensive rats. Proc. Natl. Acad. Sci. USA 1978, 75, 451–453. [Google Scholar] [CrossRef] [PubMed]
- Bateman, J.F.; Lamande, S.R.; Ramshaw, J.A. 2 Collagen Superfamily. In Extracellular Matrix, 1st ed.; Comper, W.D., Ed.; Harwood Academic Publisher: Amsterdam, The Netherlands, 1996; Volume 2, pp. 22–67. [Google Scholar]
- Von der Mark, K. Structure, Biosynthesis and Gene Regulation of Collagens in Cartilage and Bone; Academic Press: Orlando, FL, USA, 1999. [Google Scholar]
- Canty, E.G.; Kadler, K.E. Procollagen trafficking, processing and fibrillogenesis. J. Cell Sci. 2005, 118, 1341–1353. [Google Scholar] [CrossRef] [PubMed]
- Bulleid, N.J.; Dalley, J.A.; Lees, J.F. The C-propeptide domain of procollagen can be replaced with a transmembrane domain without affecting trimer formation or collagen triple helix folding during biosynthesis. EMBO J. 1997, 16, 6694–6701. [Google Scholar] [CrossRef] [PubMed]
- Lees, J.F.; Tasab, M.; Bulleid, N.J. Identification of the molecular recognition sequence which determines the type-specific assembly of procollagen. EMBO J. 1997, 16, 908–916. [Google Scholar] [CrossRef] [PubMed]
- Koivu, J. Identification of disulfide bonds in carboxy-terminal propeptides of human type I procollagen. FEBS Lett. 1987, 212, 229–232. [Google Scholar] [CrossRef]
- Bonfanti, L.; Mironov, A.A.; Martínez-Menárguez, J.A.; Martella, O.; Fusella, A.; Baldassarre, M.; Buccione, R.; Geuze, H.J.; Luini, A. Procollagen traverses the Golgi stack without leaving the lumen of cisternae: Evidence for cisternal maturation. Cell 1998, 95, 993–1003. [Google Scholar] [CrossRef]
- Stephens, D.J.; Pepperkok, R. Imaging of procollagen transport reveals COPI-dependent cargo sorting during ER-to-Golgi transport in mammalian cells. J. Cell Sci. 2002, 115, 1149–1160. [Google Scholar] [PubMed]
- Prockop, D.J.; Sieron, A.L.; Li, S.-W. Procollagen N-proteinase and procollagen C-proteinase. Two unusual metalloproteinases that are essential for procollagen processing probably have important roles in development and cell signaling. Matrix Biol. 1998, 16, 399–408. [Google Scholar] [CrossRef]
- Eyre, D.R.; Paz, M.A.; Gallop, P.M. Cross-linking in collagen and elastin. Annu. Rev. Biochem. 1984, 53, 717–748. [Google Scholar] [CrossRef] [PubMed]
- Birk, D.E.; Nurminskaya, M.V.; Zycband, E.I. Collagen fibrillogenesis in situ: Fibril segments undergo post-depositional modifications resulting in linear and lateral growth during matrix development. Dev. Dyn. 1995, 202, 229–243. [Google Scholar] [CrossRef] [PubMed]
- Lindquist, J.; Marzluff, W.; Stefanovic, B., III. Posttranscriptional regulation of type I collagen. Am. J. Physiol. 2000, 279, G471–G476. [Google Scholar]
- Lindquist, J.N.; Stekanovic, B.; Brenner, D.A. Regulation of collagen α1(I) expression in hepatic stellate cells. J. Gastroenterol. 2000, 35, 80–83. [Google Scholar] [PubMed]
- Stefanovic, B.; Hellerbrand, C.; Holcik, M.; Briendl, M.; Aliebhaber, S.; Brenner, D. Posttranscriptional regulation of collagen alpha1(I) mRNA in hepatic stellate cells. Mol. Cell. Biol. 1997, 17, 5201–5209. [Google Scholar] [CrossRef] [PubMed]
- Tsukada, S.; Parsons, C.J.; Rippe, R.A. Mechanisms of liver fibrosis. Clin. Chim. Acta 2006, 364, 33–60. [Google Scholar] [CrossRef] [PubMed]
- Määttä, A.; Ekholm, E.; Penttinen, R.P. Effect of the 3′-untranslated region on the expression levels and rnrna stability of α1(I) collagen gene. BBA Gene Struct. Expr. 1995, 1260, 294–300. [Google Scholar] [CrossRef]
- Penttinen, R.P.; Kobayashi, S.; Bornstein, P. Transforming growth factor beta increases mRNA for matrix proteins both in the presence and in the absence of changes in mRNA stability. Proc. Natl. Acad. Sci. USA 1988, 85, 1105–1108. [Google Scholar] [CrossRef] [PubMed]
- Britton, R.S.; Bacon, B.R. Intracellular signaling pathways in stellate cell activation. Alcohol. Clin. Exp. Res. 1999, 23, 922–925. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.L. Cytokines and fibrogenesis. Semin. Liver Dis. 1998, 19, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Hellerbrand, C.; Stefanovic, B.; Giordano, F.; Burchardt, E.R.; Brenner, D.A. The role of TGFβ1 in initiating hepatic stellate cell activation in vivo. J. Hepatol. 1999, 30, 77–87. [Google Scholar] [CrossRef]
- Verrecchia, F.; Mauviel, A. Transforming growth factor-beta and fibrosis. World J. Gastroenterol. 2007, 13, 3056–3062. [Google Scholar] [PubMed]
- Ihn, H. Pathogenesis of fibrosis: Role of TGF-β and CTGF. Curr. Opin. Rheumatol. 2002, 14, 681–685. [Google Scholar] [CrossRef] [PubMed]
- Ju, H.; Dixon, I.M. Effect of angiotensin II on myocardial collagen gene expression. In Biochemical Regulation of Myocardium; Springer: Berlin, Germany; Heidelberg, Germany, 1996; pp. 231–237. [Google Scholar]
- Yang, F.; Chung, A.C.; Huang, X.R.; Lan, H.Y. Angiotensin II induces connective tissue growth factor and collagen I expression via transforming growth factor-β-dependent and-independent smad pathways the role of smad3. Hypertension 2009, 54, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, J.; Lichtler, A.; Rowe, D.; Farmer, S. Cell adhesion regulates pro-alpha 1(I) collagen mRNA stability and transcription in mouse fibroblasts. J. Biol. Chem. 1991, 266, 8470–8475. [Google Scholar] [PubMed]
- Friedman, S. Hepatic stellate cells. Prog. Liver Dis. 1995, 14, 101–130. [Google Scholar]
- De Minicis, S.; Seki, E.; Uchinami, H.; Kluwe, J.; Zhang, Y.; Brenner, D.A.; Schwabe, R.F. Gene expression profiles during hepatic stellate cell activation in culture and in vivo. Gastroenterology 2007, 132, 1937–1946. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Parsons, C.J.; Stefanovic, B. Gene expression profile of quiescent and activated rat hepatic stellate cells implicates Wnt signaling pathway in activation. J. Hepatol. 2006, 45, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Rockey, D.C.; Boyles, J.; Gabbiani, G.; Friedman, S. Rat hepatic lipocytes express smooth muscle actin upon activation in vivo and in culture. J. Submicrosc. Cytol. Pathol. 1992, 24, 193–203. [Google Scholar] [PubMed]
- Cai, L.; Fritz, D.; Stefanovic, L.; Stefanovic, B. Binding of LARP6 to the conserved 5′stem–loop regulates translation of mrnas encoding type I collagen. J. Mol. Biol. 2010, 395, 309–326. [Google Scholar] [CrossRef] [PubMed]
- Stefanovic, L.; Longo, L.; Zhang, Y.; Stefanovic, B. Characterization of binding of larp6 to the 5′ stem-loop of collagen mRNAs: Implications for synthesis of type I collagen. RNA Biol. 2014, 11, 1386–1401. [Google Scholar] [CrossRef] [PubMed]
- Teplova, M.; Yuan, Y.-R.; Phan, A.T.; Malinina, L.; Ilin, S.; Teplov, A.; Patel, D.J. Structural basis for recognition and sequestration of UUU OH 3′ temini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol. Cell 2006, 21, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Kotik-Kogan, O.; Valentine, E.R.; Sanfelice, D.; Conte, M.R.; Curry, S. Structural analysis reveals conformational plasticity in the recognition of RNA 3′ ends by the human La protein. Structure 2008, 16, 852–862. [Google Scholar] [CrossRef] [PubMed]
- Bayfield, M.A.; Yang, R.; Maraia, R.J. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs). BBA Gene Regul. Mech. 2010, 1799, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Martino, L.; Pennell, S.; Kelly, G.; Busi, B.; Brown, P.; Atkinson, R.A.; Salisbury, N.J.; Ooi, Z.-H.; See, K.-W.; Smerdon, S.J.; et al. Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module. Nucleic Acids Res. 2015, 43, 645–660. [Google Scholar] [CrossRef] [PubMed]
- Bousquet-Antonelli, C.; Deragon, J.-M. A comprehensive analysis of the La-motif protein superfamily. RNA 2009, 15, 750–764. [Google Scholar] [CrossRef] [PubMed]
- Valavanis, C.; Wang, Z.; Sun, D.; Vaine, M.; Schwartz, L.M. Acheron, a novel member of the Lupus Antigen family, is induced during the programmed cell death of skeletal muscles in the moth Manduca sexta. Gene 2007, 393, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Parsons, C.J.; Stefanovic, B.; Seki, E.; Aoyama, T.; Latour, A.M.; Marzluff, W.F.; Rippe, R.A.; Brenner, D.A. Mutation of the 5′-untranslated region stem-loop structure inhibits α1(I) collagen expression in vivo. J. Biol. Chem. 2011, 286, 8609–8619. [Google Scholar] [CrossRef] [PubMed]
- Stefanovic, B.; Schnabl, B.; Brenner, D.A. Inhibition of collagen α1(I) expression by the 5′ stem-loop as a molecular decoy. J. Biol. Chem. 2002, 277, 18229–18237. [Google Scholar] [CrossRef] [PubMed]
- Holcik, M.; Liebhaber, S.A. Four highly stable eukaryotic mRNAs assemble 3′ untranslated region RNA–protein complexes sharing cis and trans components. Proc. Natl. Acad. Sci. USA 1997, 94, 2410–2414. [Google Scholar] [CrossRef] [PubMed]
- Challa, A.A.; Stefanovic, B. A novel role of vimentin filaments: Binding and stabilization of collagen mRNAs. Mol. Cell. Biol. 2011, 31, 3773–3789. [Google Scholar] [CrossRef] [PubMed]
- Galigniana, M.D.; Scruggs, J.L.; Herrington, J.; Welsh, M.J.; Carter-Su, C.; Housley, P.R.; Pratt, W.B. Heat shock protein 90-dependent (geldanamycin-inhibited) movement of the glucocorticoid receptor through the cytoplasm to the nucleus requires intact cytoskeleton. Mol. Endocrinol. 1998, 12, 1903–1913. [Google Scholar] [CrossRef] [PubMed]
- Raats, J.M.; Gerards, W.L.; Schreuder, M.I.; Grund, C.; Henderik, J.B.; Hendriks, I.; Ramaekers, F.; Bloemendal, H. Biochemical and structural aspects of transiently and stably expressed mutant desmin in vimentin-free and vimentin-containing cells. Eur. J. Cell Biol. 1992, 58, 108–127. [Google Scholar] [PubMed]
- Sjöberg, G.; Saavedra-Matiz, C.A.; Rosen, D.R.; Wijsman, E.M.; Borg, K.; Horowitz, S.H.; Sejersen, T. A missense mutation in the desmin rod domain is associated with autosomal dominant distal myopathy, and exerts a dominant negative effect on filament formation. Hum. Mol. Genet. 1999, 8, 2191–2198. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Hijikata, T.; Lin, Z.; Sweeney, H.; Englander, S.; Holtzer, H. Truncated desmin in PtK2 cells induces desmin-vimentin-cytokeratin coprecipitation, involution of intermediate filament networks, and nuclear fragmentation: A model for many degenerative diseases. Proc. Natl. Acad. Sci. USA 1994, 91, 2497–2501. [Google Scholar] [CrossRef] [PubMed]
- Kozak, M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987, 15, 8125–8148. [Google Scholar] [CrossRef] [PubMed]
- Kozak, M. An analysis of vertebrate mRNA sequences: Intimations of translational control. J. Cell Biol. 1991, 115, 887–903. [Google Scholar] [CrossRef] [PubMed]
- Hentze, M.W.; Kühn, L.C. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc. Natl. Acad. Sci. USA 1996, 93, 8175–8182. [Google Scholar] [CrossRef] [PubMed]
- Cox, T.C.; Bawden, M.J.; Martin, A.; May, B.K. Human erythroid 5-aminolevulinate synthase: Promoter analysis and identification of an iron-responsive element in the mRNA. EMBO J. 1991, 10, 1891–1902. [Google Scholar] [PubMed]
- Dandekar, T.; Stripecke, R.; Gray, N.K.; Goossen, B.; Constable, A.; Johansson, H.E.; Hentze, M.W. Identification of a novel iron-responsive element in murine and human erythroid delta-aminolevulinic acid synthase mRNA. EMBO J. 1991, 10, 1903–1909. [Google Scholar] [PubMed]
- Bhasker, C.R.; Burgiel, G.; Neupert, B.; Emery-Goodman, A.; Kühn, L.; May, B.K. The putative iron-responsive element in the human erythroid 5-aminolevulinate synthase mRNA mediates translational control. J. Biol. Chem. 1993, 268, 12699–12705. [Google Scholar] [PubMed]
- Melefors, Ö.; Goossen, B.; Johansson, H.E.; Stripecke, R.; Gray, N.K.; Hentze, M. Translational control of 5-aminolevulinate synthase mRNA by iron-responsive elements in erythroid cells. J. Biol. Chem. 1993, 268, 5974–5978. [Google Scholar] [PubMed]
- Schalinske, K.L.; Chen, O.S.; Eisenstein, R.S. Iron differentially stimulates translation of mitochondrial aconitase and ferritin mRNAs in mammalian cells implications for iron regulatory proteins as regulators of mitochondrial citrate utilization. J. Biol. Chem. 1998, 273, 3740–3746. [Google Scholar] [CrossRef] [PubMed]
- Kohler, S.A.; Henderson, B.R.; Kühn, L.C. Succinate dehydrogenase b mRNA of drosophila melanogaster has a functional iron-responsive element in its 5′-untranslated region. J. Biol. Chem. 1995, 270, 30781–30786. [Google Scholar] [CrossRef] [PubMed]
- Akopian, D.; Shen, K.; Zhang, X.; Shan, S.-O. Signal recognition particle: An essential protein targeting machine. Annu. Rev. Biochem. 2013, 82, 693–721. [Google Scholar] [CrossRef] [PubMed]
- Nicchitta, C.V.; Lerner, R.S.; Stephens, S.B.; Dodd, R.D.; Pyhtila, B. Pathways for compartmentalizing protein synthesis in eukaryotic cells: The template-partitioning model. Biochem. Cell Biol. 2005, 83, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Nicchitta, C.V. A platform for compartmentalized protein synthesis: Protein translation and translocation in the ER. Curr. Opin. Cell Biol. 2002, 14, 412–416. [Google Scholar] [CrossRef]
- Hermesh, O.; Jansen, R.-P. Take the (RN)A-train: Localization of mRNA to the endoplasmic reticulum. BBA Mol. Cell Res. 2013, 1833, 2519–2525. [Google Scholar] [CrossRef] [PubMed]
- Kraut-Cohen, J.; Afanasieva, E.; Haim-Vilmovsky, L.; Slobodin, B.; Yosef, I.; Bibi, E.; Gerst, J.E. Translation-and SRP-independent mRNA targeting to the endoplasmic reticulum in the yeast saccharomyces cerevisiae. Mol. Biol. Cell 2013, 24, 3069–3084. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.A.; Zhang, H.; Palazzo, A.F. P180 promotes the ribosome-independent localization of a subset of mRNA to the endoplasmic reticulum. PLoS Biol. 2012, 10, e1001336. [Google Scholar] [CrossRef] [PubMed]
- Pyhtila, B.; Zheng, T.; Lager, P.J.; Keene, J.D.; Reedy, M.C.; Nicchitta, C.V. Signal sequence-and translation-independent mRNA localization to the endoplasmic reticulum. RNA 2008, 14, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Stefanovic, B. Role of LARP6 and nonmuscle myosin in partitioning of collagen mRNAs to the ER membrane. PLoS ONE 2014, 9, e108870. [Google Scholar] [CrossRef] [PubMed]
- Stefanovic, B.; Stefanovic, L.; Schnabl, B.; Bataller, R.; Brenner, D.A. TRAM2 protein interacts with endoplasmic reticulum Ca2+ pump serca2b and is necessary for collagen type I synthesis. Mol. Cell. Biol. 2004, 24, 1758–1768. [Google Scholar] [CrossRef] [PubMed]
- Löfgren, M.; Ekblad, E.; Morano, I.; Arner, A. Nonmuscle myosin motor of smooth muscle. J. Gen. Physiol. 2003, 121, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Simerly, C.; Nowak, G.; de Lanerolle, P.; Schatten, G. Differential expression and functions of cortical myosin IIA and IIB isotypes during meiotic maturation, fertilization, and mitosis in mouse oocytes and embryos. Mol. Biol. Cell 1998, 9, 2509–2525. [Google Scholar] [CrossRef] [PubMed]
- Isemura, M.; Mita, T.; Satoh, K.; Narumi, K.; Motomiya, M. Myosin light chain kinase inhibitors ML-7 and ML-9 inhibit mouse lung carcinoma cell attachment to the fibronectin substratum. Cell Biol. Int. Rep. 1991, 15, 965–972. [Google Scholar] [CrossRef]
- Connell, L.E.; Helfman, D.M. Myosin light chain kinase plays a role in the regulation of epithelial cell survival. J. Cell Sci. 2006, 119, 2269–2281. [Google Scholar] [CrossRef] [PubMed]
- Kovács, M.; Tóth, J.; Hetényi, C.; Málnási-Csizmadia, A.; Sellers, J.R. Mechanism of blebbistatin inhibition of myosin II. J. Biol. Chem. 2004, 279, 35557–35563. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Fritz, D.; Stefanovic, L.; Stefanovic, B. Nonmuscle myosin-dependent synthesis of type I collagen. J. Mol. Biol. 2010, 401, 564–578. [Google Scholar] [CrossRef] [PubMed]
- Tangkijvanich, P.; Tam, S.P.; Yee, H.F. Wound-induced migration of rat hepatic stellate cells is modulated by endothelin-1 through rho-kinase-mediated alterations in the acto-myosin cytoskeleton. Hepatology 2001, 33, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Pandya, K.; Kim, H.-S.; Smithies, O. Fibrosis, not cell size, delineates β-myosin heavy chain reexpression during cardiac hypertrophy and normal aging in vivo. Proc. Natl. Acad. Sci. USA 2006, 103, 16864–16869. [Google Scholar] [CrossRef] [PubMed]
- Hunt, S.L.; Hsuan, J.J.; Totty, N.; Jackson, R.J. Unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev. 1999, 13, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, C.; Usui, K.; Ito, F.; Itoh, M.; Hayashizaki, Y.; Suzuki, H. Role of survival motor neuron complex components in small nuclear ribonucleoprotein assembly. J. Biol. Chem. 2009, 284, 14609–14617. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.V.; Delrow, J.; Corrin, P.D.; Frazier, J.P.; Soriano, P. Identification and validation of PDGF transcriptional targets by microarray-coupled gene-trap mutagenesis. Nat. Genet. 2004, 36, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Datta, P.K.; Chytil, A.; Gorska, A.E.; Moses, H.L. Identification of STRAP, a novel WD domain protein in transforming growth factor-β signaling. J. Biol. Chem. 1998, 273, 34671–34674. [Google Scholar] [CrossRef] [PubMed]
- Margottin, F.; Bour, S.P.; Durand, H.; Selig, L.; Benichou, S.; Richard, V.; Thomas, D.; Strebel, K.; Benarous, R. A novel human WD protein, h-βTrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell 1998, 1, 565–574. [Google Scholar] [CrossRef]
- Komachi, K.; Redd, M.J.; Johnson, A.D. The WD repeats of Tup1 interact with the homeo domain protein alpha 2. Genes Dev. 1994, 8, 2857–2867. [Google Scholar] [CrossRef] [PubMed]
- Vukmirovic, M.; Manojlovic, Z.; Stefanovic, B. Serine-threonine kinase receptor-associated protein (STRAP) regulates translation of type I collagen mRNAs. Mol. Cell. Biol. 2013, 33, 3893–3906. [Google Scholar] [CrossRef] [PubMed]
- Merret, R.; Martino, L.; Bousquet-Antonelli, C.; Fneich, S.; Descombin, J.; Billey, É.; Conte, M.R.; Deragon, J.-M. The association of a La module with the PABP-interacting motif PAM2 is a recurrent evolutionary process that led to the neofunctionalization of La-related proteins. RNA 2013, 19, 36–50. [Google Scholar] [CrossRef] [PubMed]
- Manojlovic, Z.; Stefanovic, B. A novel role of RNA helicase A in regulation of translation of type I collagen mRNAs. RNA 2012, 18, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Grosse, F. Multiple functions of nuclear DNA helicase II (RNA helicase A) in nucleic acid metabolism. Acta Biochim. Biophys. Sin. 2004, 36, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Hartman, T.R.; Qian, S.; Bolinger, C.; Fernandez, S.; Schoenberg, D.R.; Boris-Lawrie, K. RNA helicase A is necessary for translation of selected messenger RNAs. Nat. Struct. Mol. Biol. 2006, 13, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Fujii, R.; Okamoto, M.; Aratani, S.; Oishi, T.; Ohshima, T.; Taira, K.; Baba, M.; Fukamizu, A.; Nakajima, T. A role of RNA helicase A in cis-acting transactivation response element-mediated transcriptional regulation of human immunodeficiency virus type 1. J. Biol. Chem. 2001, 276, 5445–5451. [Google Scholar] [CrossRef] [PubMed]
- Bolinger, C.; Yilmaz, A.; Hartman, T.R.; Kovacic, M.B.; Fernandez, S.; Ye, J.; Forget, M.; Green, P.L.; Boris-Lawrie, K. RNA helicase A interacts with divergent lymphotropic retroviruses and promotes translation of human T-cell leukemia virus type 1. Nucleic Acids Res. 2007, 35, 2629–2642. [Google Scholar] [CrossRef] [PubMed]
- Butsch, M.; Hull, S.; Wang, Y.; Roberts, T.M.; Boris-Lawrie, K. The 5′ RNA terminus of spleen necrosis virus contains a novel posttranscriptional control element that facilitates human immunodeficiency virus Rev/RRE-independent gag production. J. Virol. 1999, 73, 4847–4855. [Google Scholar] [PubMed]
- Manojlovic, Z.; Blackmon, J.; Stefanovic, B. Tacrolimus (FK506) prevents early stages of ethanol induced hepatic fibrosis by targeting LARP6 dependent mechanism of collagen synthesis. PLoS ONE 2013, 8, e65897. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.J.; Burakoff, S.J. The 25-kDa FK506-binding protein is localized in the nucleus and associates with casein kinase II and nucleolin. Proc. Natl. Acad. Sci. USA 1993, 90, 7769–7773. [Google Scholar] [CrossRef] [PubMed]
- Ochocka, A.M.; Kampanis, P.; Nicol, S.; Allende-Vega, N.; Cox, M.; Marcar, L.; Milne, D.; Fuller-Pace, F.; Meek, D. FKBP25, a novel regulator of the p53 pathway, induces the degradation of MDM2 and activation of p53. FEBS Lett. 2009, 583, 621–626. [Google Scholar] [CrossRef] [PubMed]
- Bram, R.J.; Hung, D.T.; Martin, P.K.; Schreiber, S.L.; Crabtree, G.R. Identification of the immunophilins capable of mediating inhibition of signal transduction by cyclosporin A and FK506: Roles of calcineurin binding and cellular location. Mol. Cell. Biol. 1993, 13, 4760–4769. [Google Scholar] [CrossRef] [PubMed]
- Gudavicius, G.; Dilworth, D.; Serpa, J.J.; Sessler, N.; Petrotchenko, E.V.; Borchers, C.H.; Nelson, C.J. The prolyl isomerase, FKBP25, interacts with RNA-engaged nucleolin and the pre-60S ribosomal subunit. RNA 2014, 20, 1014–1022. [Google Scholar] [CrossRef] [PubMed]
- Galat, A. Peptidylproline cis-trans-isomerases: Immunophilins. Eur. J. Biochem. 1993, 216, 689–707. [Google Scholar] [CrossRef] [PubMed]
- Nagano, J.; Iyonaga, K.; Kawamura, K.; Yamashita, A.; Ichiyasu, H.; Okamoto, T.; Suga, M.; Sasaki, Y.; Kohrogi, H. Use of tacrolimus, a potent antifibrotic agent, in bleomycin-induced lung fibrosis. Eur. Respir. J. 2006, 27, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Stefanovic, B. Akt mediated phosphorylation of LARP6; critical step in biosynthesis of type I collagen. Sci. Rep. 2016, 6, 22597. [Google Scholar] [CrossRef] [PubMed]


© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Stefanovic, B. LARP6 Meets Collagen mRNA: Specific Regulation of Type I Collagen Expression. Int. J. Mol. Sci. 2016, 17, 419. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17030419
Zhang Y, Stefanovic B. LARP6 Meets Collagen mRNA: Specific Regulation of Type I Collagen Expression. International Journal of Molecular Sciences. 2016; 17(3):419. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17030419
Chicago/Turabian StyleZhang, Yujie, and Branko Stefanovic. 2016. "LARP6 Meets Collagen mRNA: Specific Regulation of Type I Collagen Expression" International Journal of Molecular Sciences 17, no. 3: 419. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17030419