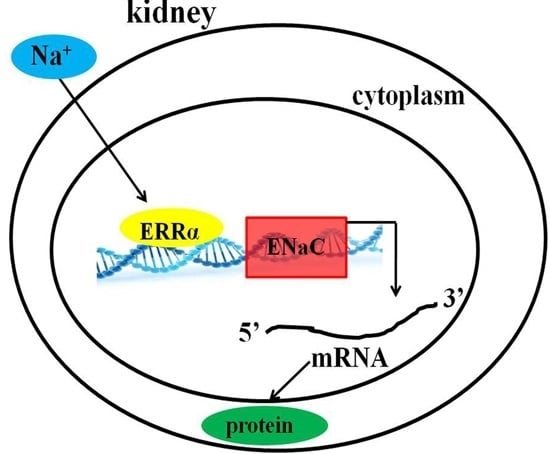
High Salt Diet Affects Renal Sodium Excretion and ERRα Expression
Abstract
:1. Introduction
2. Results
2.1. Effects of Salt Loading on Systolic Blood Pressure and Weight
2.2. Characteristic Response to Dietary and Drug Intervention
2.3. Impact of Different Interventions on the Expression of ERRα in the Kidneys of SS.13BN and Dahl Salt-Sensitive (DS) Rats
2.4. The mRNA and Protein Expressions of Na+/K+-ATPase and Epithelial Sodium Channel (ENaC)
2.5. Chromatin Immunoprecipitation Assay (CHIP)
3. Discussion
4. Materials and Methods
4.1. Experimental Animals and Protocols
4.2. Measurement of Blood Pressure and Body Weight
4.3. Measurement of Blood and Urine Biochemical Indicators
4.4. Immunohistochemistry
4.5. QRT-PCR for ERRα, Na+/K+-ATPase and α-, β-, γ-ENaC Expressions in Kidneys
4.6. Western Blot Analyses of ERRα, Na+/K+-ATPase, and β-, γ-ENaC Expressions in Kidneys
4.7. Chromatin Immunoprecipitation (CHIP) Assay
4.8. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sullivan, J.M. Salt sensitivity: Definition, conception, methodology, and long-term issues. Hypertension 1991, 17, 61–68. [Google Scholar] [CrossRef]
- Dahl, L.K.; Heine, M. Primary role of renal homografts in setting chronic blood pressure levels in rats. Circ. Res. 1975, 36, 692–696. [Google Scholar] [CrossRef] [PubMed]
- Rettig, R. Does the kidney play a role in the aetiology of primary hypertension? Evidence from renal transplantation studies in rats and humans. J. Hum. Hypertens. 1993, 7, 177–180. [Google Scholar] [PubMed]
- Kakizoe, Y.; Kitamura, K.; Ko, T.; Wakida, N.; Maekawa, A.; Miyoshi, T.; Shiraishi, N.; Adachi, M.; Zhang, Z.; Masilamani, S.; et al. Aberrant ENaC activation in Dahl salt-sensitive rats. J. Hypertens. 2009, 27, 1679–1689. [Google Scholar] [CrossRef] [PubMed]
- Shehata, M.F. Regulation of the epithelial sodium channel [ENaC] in kidneys of salt-sensitive Dahl rats: Insights on alternative splicing. Int. Arch. Med. 2009, 2, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Periyasamy, S.M.; Liu, J.; Tanta, F.; Kabak, B.; Wakefield, B.; Malhotra, D.; Kennedy, D.J.; Nadoor, A.; Fedorova, O.V.; Gunning, W.; et al. Salt loading induces redistribution of the plasmalemmal Na/K-ATPase in proximal tubule cells. Kidney Int. 2005, 67, 1868–1877. [Google Scholar] [CrossRef] [PubMed]
- Sanada, H.; Jones, J.E.; Jose, P.A. Genetics of salt-sensitive hypertension. Curr. Hypertens. Rep. 2010, 13, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Giguere, V.; Yang, N.; Segui, P.; Evans, R.M. Identification of a new class of steroid hormone receptors. Nature 1988, 331, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Sladek, R.; Bader, J.A.; Giguere, V. The orphan nuclear receptor estrogenrelated receptor α is a transcriptional regulator of the human medium-chain acyl coenzyme A dehydrogenase gene. Mol. Cell. Biol. 1997, 17, 5400–5409. [Google Scholar] [CrossRef] [PubMed]
- Cavallini, A.; Notarnicola, M.; Giannini, R.; Montemurro, S.; Lorusso, D.; Visconti, A.; Minervini, F.; Caruso, M.G. Oestrogen receptor-related receptor α (ERRα) and oestrogen receptors (ERα and ERβ) exhibit different gene expression in human colorectal tumour progression. Eur. J. Cancer 2005, 41, 1487–1494. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Sladek, R.; Carrier, J.; Bader, J.A.; Richard, D.; Giguère, V. Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor α. Mol. Cell. Biol. 2003, 23, 7947–7956. [Google Scholar] [CrossRef] [PubMed]
- Huss, J.M.; Torra, I.P.; Staels, B.; Giguère, V.; Kelly, D.P. Estrogen-related receptor α directs peroxisome proliferator-activated receptor a signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol. Cell. Biol. 2004, 24, 9079–9091. [Google Scholar] [CrossRef] [PubMed]
- Carrier, J.C.; Deblois, G.; Champigny, C.; Levy, E.; Giguère, V. Estrogen-related receptor α (ERRα) is a transcriptional regulator of apolipoprotein A-IV and controls lipid handling in the intestine. J. Biol. Chem. 2004, 279, 52052–52058. [Google Scholar] [CrossRef] [PubMed]
- Alaynick, W.A.; Way, J.M.; Wilson, S.A.; Benson, W.G.; Pei, L.; Downes, M.; Yu, R.; Jonker, J.W.; Holt, J.A.; Rajpal, D.K.; et al. ERRγ regulates cardiac, gastric, and renal potassium homeostasis. Mol. Endocrinol. 2010, 24, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, A.M.; Dufour, C.R.; Ghahremani, M.; Reudelhuber, T.L.; Giguère, V. Physiological genomics identifies estrogen-related receptor α as a regulator of renal sodium and potassium homeostasis and the renin-angiotensin pathway. Mol. Endocrinol. 2010, 24, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Giguere, V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr. Rev. 2008, 29, 677–696. [Google Scholar] [CrossRef] [PubMed]
- Chiolero, A.W.; Grgoire, B.; Burnier, M. Renal determinants of the salt sensitivity of blood pressure. Nephrol. Dial. Transplant. 2001, 16, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Rapp, J.P.; Dene, H. Development and characteristics of inbred strains of Dahl salt-sensitive and salt-resistant rats. Hypertension 1985, 7, 340–349. [Google Scholar] [PubMed]
- Fujita, T. Mineralocorticoid receptors, salt-sensitive hypertension, and metabolic syndrome. Hypertension 2010, 55, 813–818. [Google Scholar] [CrossRef] [PubMed]
- Weinberger, M.H.; Fineberg, N.S.; Fineberg, S.E.; Weinberger, M. Salt sensitivity, pulse pressure, and death in normal and hypertensive humans. Hypertension 2001, 37, 429–432. [Google Scholar] [CrossRef] [PubMed]
- Coffman, T.M.; Crowley, S.D. Kidney in hypertension: Guyton redux. Hypertension 2008, 51, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Guyton, A.C.; Coleman, T.G.; Cowley, A.V.J.; Scheel, K.W.; Manning, R.D., Jr.; Norman, R.A., Jr. Arterial pressure regulation. Overriding dominance of the kidneys in long-term regulation and in hypertension. Am. J. Med. 1972, 52, 584–594. [Google Scholar] [CrossRef]
- Johnson, R.J.; Herrera-Acosta, J.; Schreiner, G.F.; Rodriguez-Iturbe, B. Subtle acquired renal injury as a mechanism of salt-sensitive hypertension. N. Engl. J. Med. 2002, 346, 913–923. [Google Scholar] [CrossRef] [PubMed]
- Ideker, T.; Thorsson, V.; Ranish, J.A.; Christmas, R.; Buhler, J.; Eng, J.K.; Bumgarner, R.; Goodlett, D.R.; Aebersold, R.; Hood, L. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 2001, 292, 929–934. [Google Scholar] [CrossRef] [PubMed]
- Aoi, W.; Niisato, N.; Sawabe, Y.; Miyazaki, H.; Tokuda, S.; Nishio, K.; Yoshikawa, T.; Marunaka, Y. Abnormal expression of ENaC and SGK1 mRNA induced by dietary sodium in Dahl salt-sensitively hypertensive rats. Cell Biol. Int. 2007, 31, 1288–1291. [Google Scholar] [CrossRef] [PubMed]
- Berman, J.M.; Brand, C.; Awayda, M.S. A long isoform of the epithelial sodium channel α subunit forms a highly active channel. Channels 2015, 9, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Haloui, M.; Tremblay, J.; Seda, O.; Koltsova, S.V.; Maksimov, G.V.; Orlov, S.N.; Hamet, P. Increased renal epithelial Na channel expression and activity correlate with elevation of blood pressure in spontaneously hypertensive rats. Hypertension 2013, 62, 731–737. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.S.; Warnock, D.G. Disorders of the epithelial Na+ Channel in Liddle’s syndrome and autosomal recessive pseudohypoaldosteronism type 1. Exp. Nephrol. 2000, 8, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Jaitovich, A.; Bertorello, A.M. Salt, Na+, K+-ATPase and hypertension. Life Sci. 2010, 86, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Bagrov, A.Y.; Agalakova, N.I.; Kashkin, V.A.; Fedorova, O.V. Endogenous cardiotonic steroids and differential patterns of sodium pump inhibition in NaCl-loaded salt-sensitive and normotensive rats. Am. J. Hypertens. 2009, 22, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Fedorova, O.V.; Talan, M.I.; Agalakova, N.I.; Lakatta, E.G.; Bagrov, A.Y. Coordinated shifts in Na/K-ATPase isoforms and their endogenous ligands during cardiac hypertrophy and failure in NaCl-sensitive hypertension. J. Hypertens. 2004, 22, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Fedorova, O.V.; Agalakova, N.I.; Talan, M.I.; Lakatta, E.G.; Bagrov, A.Y. Brain ouabain stimulates peripheral marinobufagenin via angiotensin II signalling in NaCl-loaded Dahl-S rats. J. Hypertens. 2005, 23, 1515–1523. [Google Scholar] [CrossRef] [PubMed]
- Ariazi, E.A.; Jordan, V.C. Estrogen-related receptors as emerging targets in cancer and metabolic disorders. Curr. Top. Med. Chem. 2006, 6, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Huss, J.M.; Imahashi, K.I.; Dufour, C.R.; Weinheimer, C.J.; Courtois, M.; Kovacs, A.; Giguère, V.; Murphy, E.; Kelly, D.P. The nuclear receptor ERRα is required for the bioenergetic and functional adaptation to cardiac pressure overload. Cell Metab. 2007, 6, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.; Yang, L.; Stallcup, M.R. Hormone-independent transcriptional activation and coactivator binding by novel orphan nuclear receptor ERR3. J. Biol. Chem. 1999, 274, 22618–22626. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Xu, X.; Lu, Z.; Zhang, P.; Fassett, J.; Zhang, Y.; Xin, Y.; Hall, J.L.; Viollet, B.; Bache, R.J.; et al. AMP activated protein kinase-α2 regulates expression of estrogen-related receptor-α, a metabolic transcription factor related to heart failure development. Hypertension 2011, 58, 696–703. [Google Scholar] [CrossRef] [PubMed]
- Lee, V.H.; Lam, I.P.; Choi, H.S.; Chow, B.K.; Lee, L.T. The estrogen-related receptor α upregulates secretin expressions in response to hypertonicity and angiotensin II stimulation. PLoS ONE 2012, 7, e39913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivera, P.; Pérez-Martín, M.; Pavón, F.J.; Crespillo, A.; Cifuentes, M.; López-Ávalos, M.D.; Grondona, J.M.; Vida, M.; Fernández-Llebrez, P.; de Fonseca, F.R.; et al. Pharmacological administration of the isoflavone daidzein enhances cell proliferation and reduces high fat diet-induced apoptosis and gliosis in the rat hippocampus. PLoS ONE 2013, 8, e64750. [Google Scholar]
- Nelson, J.D.; Denisenko, O.; Bomsztyk, K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 2006, 1, 179–185. [Google Scholar] [CrossRef] [PubMed]




| Group | Weight (g) | SBP (mm Hg) | ||
|---|---|---|---|---|
| Baseline | Intervention | Baseline | Intervention | |
| BN | 266.00 ± 5.18 | 340.60 ± 2.82 | 123.57 ± 1.67 | 144.71 ± 2.15 |
| BH | 262.63 ± 4.90 | 309.13 ± 6.05 # | 122.14 ± 2.61 | 146.43 ± 3.17 |
| BHD | 262.83 ± 1.92 | 301.00 ± 1.77 | 120.67 ± 1.20 | 147.67 ± 1.80 |
| SN | 268.17 ± 3.87 | 344.83 ± 8.64 | 121.29 ± 2.60 | 146.14 ± 2.88 |
| SH | 273.86 ± 5.57 | 313.4 ± 5.63 ** | 125.00 ± 3.00 | 174.71 ± 2.12 * |
| SHD | 263.00 ± 2.86 | 306.56 ± 4.04 | 125.43 ± 2.03 | 155.43 ± 2.93 |
| Group | Baseline | 1 Week | 2 Weeks | 3 Weeks | 4 Weeks | 5 Weeks | 6 Weeks |
|---|---|---|---|---|---|---|---|
| BN | 123.57 ± 1.67 | 133.00 ± 2.42 | 135.86 ± 2.21 | 137.14 ± 2.65 | 131.29 ± 1.76 | 136.71 ± 3.03 | 144.71 ± 2.15 |
| BH | 122.14 ± 2.61 | 136.29 ± 1.84 | 140.71 ± 2.42 | 142.14 ± 2.20 | 140.71 ± 1.85 # | 144.29 ± 2.34 # | 146.43 ± 3.17 |
| BHD | 120.67 ± 1.20 | 125.67 ± 1.63 | 131.50 ± 0.85 | 134.67 ± 1.56 | 143.00 ± 2.08 | 142.67 ± 2.16 | 147.67 ± 1.80 |
| SN | 121.29 ± 3.00 | 132.86 ± 2.76 | 146.57 ± 2.09 | 144.71 ± 3.23 | 145.86 ± 2.30 | 145.29 ± 3.02 | 146.14 ± 2.88 |
| SH | 125.00 ± 3.00 | 142.86 ± 2.11 * | 150.71 ± 2.67 $ | 167.86 ± 3.07 * | 161.29 ± 1.91 * | 166.43 ± 3.29 * | 174.71 ± 2.12 * |
| SHD | 125.43 ± 2.03 | 132.29 ± 2.30 | 145.57 ± 2.63 & | 152.00 ± 2.73 & | 152.00 ± 2.82 & | 158.86 ± 1.10 & | 155.43 ± 2.93 & |
| Parameters | BN | BH | BHD | SN | SH | SHD |
|---|---|---|---|---|---|---|
| Left kidney weight (g) | 1.16 ± 0.03 | 1.21 ± 0.04 | 1.17 ± 0.04 | 1.15 ± 0.02 | 1.28 ± 0.03 ** | 1.2 ± 0.08 |
| KW/BW (‰) | 3.40 ± 0.09 | 3.80 ± 0.10 ## | 3.84 ± 0.13 | 3.44 ± 0.07 | 4.36 ± 0.11 **,$ | 4.05 ± 0.23 |
| Blood parameters (μmol/L) | ||||||
| Serum creatinine | 50.13 ± 3.16 | 47.93 ± 2.36 | 46.28 ± 0.46 | 53.35 ± 3.47 | 43.55 ± 2.12 ** | 48.33 ± 1.27 |
| Serum sodium | 137.35 ± 0.43 | 139.40 ± 0.90 # | 136.13 ± 0.14 | 138.80 ± 0.36 | 140.53 ± 0.68 ** | 138.88 ± 0.60 |
| Serum potassium | 4.50 ± 0.04 | 4.62 ± 0.04 | 4.65 ± 0.13 | 4.44 ± 0.05 | 4.37 ± 0.04 $ | 4.86 ± 0.04 |
| Urinary parameters | ||||||
| 24 h urinary sodium (mmol/day) | 3.86 ± 0.19 | 41.15 ± 0.80 ## | 55.46 ± 2.32 | 3.34 ± 0.62 | 28.73 ± 1.59 * | 39.00 ± 0.82 & |
| 24 h urinary potassium (mmol/day) | 1.81 ± 0.07 | 5.57 ± 0.30 ## | 3.21 ± 0.43 | 0.97 ± 0.05 | 9.44 ± 0.53 * | 6.12 ± 0.40 & |
| 24 h urinary albumin (mg/day) | 24.19 ± 3.10 | 195.63 ± 34.00 ## | 182.24 ± 23.10 | 17.25 ± 6.43 | 234.85 ± 27.00 ** | 186.52 ± 30.41 |
| 24 h urine volume (mL) | 9.00 ± 0.71 | 62.25 ± 5.17 ## | 73.50 ± 3.62 | 8.00 ± 0.41 | 52.25 ± 5.84 && | 65.75 ± 5.90 |
| Microalbuminuria (mg/day) | 0.71 ± 0.04 | 4.97 ± 0.23 ## | 4.58 ± 0.73 | 0.46 ± 0.15 | 6.05 ± 0.61 ** | 5.65 ± 0.82 |
| Creatinine clearance (mL/min) | 0.83 ± 0.12 | 0.72 ± 0.11 | 0.82 ± 0.07 | 0.80 ± 0.07 | 0.43 ± 0.07 * | 0.70 ± 0.05 |
| Gene | Primers | Sequence (5′-3′) | Primer Length (bp) |
|---|---|---|---|
| GAPDH | Forward primer | GGCACAGTCAAGGCTGAGAATG | 22 |
| Reverse primer | ATGGTGGTGAAGACGCCAGTA | 21 | |
| ERRα | Forward primer | GATGTGGCCTCTGGCTACCACTA | 23 |
| Reverse primer | CGGACAGCTGTACTCGATGCTC | 22 | |
| Na-K-ATPase | Forward primer | ACTGGCAGAGAACGGCTTCCT | 21 |
| Reverse primer | TCCGCTGCTCATAGGTCCACTC | 22 | |
| α-ENaC | Forward primer | TGGTACCGCTTCCATTACAT | 20 |
| Reverse primer | AGCGACAGGTGAAGATGAAG | 20 | |
| β-ENaC | Forward primer | CACACCAACTGTGTCTTCCA | 20 |
| Reverse primer | CATAGTTGGGAGGTGGAGTG | 20 | |
| γ-ENaC | Forward primer | TCCTTGTATGGGGTCAAAGA | 20 |
| Reverse primer | GAGCACAATTGAAATCCCAC | 20 | |
| c-Atp1a3-1 | Forward primer | TCACTGAACACGCCTGCTAC | 20 |
| Reverse primer | TCCTGCCTTCTGACTGGGAT | 20 | |
| c-Atp1a3-2 | Forward primer | GTACCACGAGCCTTGTCTCAT | 21 |
| Reverse primer | TTCAACCAGAAGCTCAAAAACCAG | 24 | |
| c-ENaC-1 | Forward primer | CCAGCCCCTACTTCACCTG | 19 |
| Reverse primer | GAAAAGAGAGACAGACTGACAGGAC | 25 | |
| c-ENaC-2 | Forward primer | GGAGCCAGTCAAACAGTCCG | 20 |
| Reverse primer | CATGGACCCCTTAGGCGAG | 19 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Wang, Y.; Liu, F.-Q.; Yuan, Z.-Y.; Mu, J.-J. High Salt Diet Affects Renal Sodium Excretion and ERRα Expression. Int. J. Mol. Sci. 2016, 17, 480. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17040480
Wang D, Wang Y, Liu F-Q, Yuan Z-Y, Mu J-J. High Salt Diet Affects Renal Sodium Excretion and ERRα Expression. International Journal of Molecular Sciences. 2016; 17(4):480. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17040480
Chicago/Turabian StyleWang, Dan, Yang Wang, Fu-Qiang Liu, Zu-Yi Yuan, and Jian-Jun Mu. 2016. "High Salt Diet Affects Renal Sodium Excretion and ERRα Expression" International Journal of Molecular Sciences 17, no. 4: 480. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17040480