Proteome Profile and Quantitative Proteomic Analysis of Buffalo (Bubalusbubalis) Follicular Fluid during Follicle Development
Abstract
:1. Introduction
2. Results
2.1. Proteome Profile of BFF
2.2. Gene Ontology (GO) Analysis
2.3. KEGG Pathway Analysis
2.4. TMT-Labeled Quantitative Analysis
2.5. Western Blot Analysis of Vimentin
3. Discussion
3.1. Profile of BFF Proteome
3.2. Comparison of BFF, HFF and Plasma
3.3. Protein Functional Classification and Mining
3.4. Extracellular Matrix (ECM) Proteins and Signaling Proteins
3.5. Protein–Protein Interaction Network
3.6. Quantitative Proteomic Analysis
4. Materials and Methods
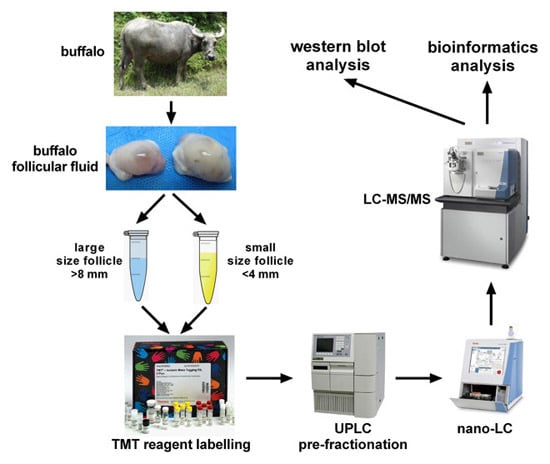
4.1. Follicular Fluid Collection and Preparation
4.2. SDS-PAGE and LC-MS/MS Analysis
4.3. Protein In-Solution Digestion and Peptide Labeling
4.4. Peptide Pre-Fractionation and LC–MS/MS Analysis
4.5. Database Searching and Bioinformatics Analysis
4.6. Western Blot Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fortune, J.E.; Rivera, G.M.; Yang, M.Y. Follicular development: The role of the follicular microenvironment in selection of the dominant follicle. Anim. Reprod. Sci. 2004, 82–83, 109–126. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, R.J.; Irving-Rodgers, H.F. Formation of the ovarian follicular antrum and follicular fluid. Biol. Reprod. 2010, 82, 1021–1029. [Google Scholar] [CrossRef] [PubMed]
- Fortune, J.E. Ovarian follicular growth and development in mammals. Biol. Reprod. 1994, 50, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Morado, S.A.; Cetica, P.D.; Beconi, M.T.; Dalvit, G.C. Reactive oxygen species in bovine oocyte maturation in vitro. Reprod. Fertil. Dev. 2009, 21, 608–614. [Google Scholar] [CrossRef] [PubMed]
- Richards, J.S. Hormonal control of gene expression in the ovary. Endocr. Rev. 1994, 15, 725–751. [Google Scholar] [CrossRef] [PubMed]
- Twigt, J.; Steegers-Theunissen, R.P.; Bezstarosti, K.; Demmers, J.A. Proteomic analysis of the microenvironment of developing oocytes. Proteomics 2012, 12, 1463–1471. [Google Scholar] [CrossRef] [PubMed]
- Boxmeer, J.C.; Macklon, N.S.; Lindemans, J.; Beckers, N.G.; Eijkemans, M.J.; Laven, J.S.; Steegers, E.A.; Steegers-Theunissen, R.P. Ivf outcomes are associated with biomarkers of the homocysteine pathway in monofollicular fluid. Hum. Reprod. 2009, 24, 1059–1066. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, M.; Zamah, A.M.; Conti, M. Epidermal growth factor-like growth factors in the follicular fluid: Role in oocyte development and maturation. Semin. Reprod. Med. 2009, 27, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Kim, M.S.; Lee, S.H.; Choi, B.C.; Lim, J.M.; Cha, K.Y.; Baek, K.H. Proteomic analysis of recurrent spontaneous abortion: Identification of an inadequately expressed set of proteins in human follicular fluid. Proteomics 2006, 6, 3445–3454. [Google Scholar] [CrossRef] [PubMed]
- Spitzer, D.; Murach, K.F.; Lottspeich, F.; Staudach, A.; Illmensee, K. Different protein patterns derived from follicular fluid of mature and immature human follicles. Hum. Reprod. 1996, 11, 798–807. [Google Scholar] [CrossRef] [PubMed]
- Anahory, T.; Dechaud, H.; Bennes, R.; Marin, P.; Lamb, N.J.; Laoudj, D. Identification of new proteins in follicular fluid of mature human follicles. Electrophoresis 2002, 23, 1197–1202. [Google Scholar] [CrossRef]
- Angelucci, S.; Ciavardelli, D.; di Giuseppe, F.; Eleuterio, E.; Sulpizio, M.; Tiboni, G.M.; Giampietro, F.; Palumbo, P.; di Ilio, C. Proteome analysis of human follicular fluid. Biochim. Biophys. Acta 2006, 1764, 1775–1785. [Google Scholar] [CrossRef] [PubMed]
- Hanrieder, J.; Nyakas, A.; Naessen, T.; Bergquist, J. Proteomic analysis of human follicular fluid using an alternative bottom-up approach. J. Proteome Res. 2008, 7, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.X.; Zhu, Y.M.; Luo, Q.; Wu, Y.T.; Gao, H.J.; Zhu, X.M.; Xu, C.M.; Huang, H.F. Specific peptide patterns of follicular fluids at different growth stages analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Biochim. Biophys. Acta 2007, 1770, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.C.; Lee, S.W.; Lee, K.W.; Cha, K.Y.; Kim, K.H.; Lee, S. Identification of new proteins in follicular fluid from mature human follicles by direct sample rehydration method of two-dimensional polyacrylamide gel electrophoresis. J. Korean Med. Sci. 2005, 20, 456–460. [Google Scholar] [CrossRef] [PubMed]
- Ambekar, A.S.; Nirujogi, R.S.; Srikanth, S.M.; Chavan, S.; Kelkar, D.S.; Hinduja, I.; Zaveri, K.; Prasad, T.S.; Harsha, H.C.; Pandey, A.; et al. Proteomic analysis of human follicular fluid: A new perspective towards understanding folliculogenesis. J. Proteom. 2013, 87, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Mortarino, M.; Vigo, D.; Maffeo, G.; Ronchi, S. Two-dimensional polyacrylamide gel electrophoresis map of bovine ovarian fluid proteins. Electrophoresis 1999, 20, 866–869. [Google Scholar] [CrossRef]
- Fahiminiya, S.; Labas, V.; Roche, S.; Dacheux, J.L.; Gerard, N. Proteomic analysis of mare follicular fluid during late follicle development. Proteome Sci. 2011, 9, 54. [Google Scholar] [CrossRef] [PubMed]
- Bijttebier, J.; Tilleman, K.; Dhaenens, M.; Deforce, D.; van Soom, A.; Maes, D. Comparative proteome analysis of porcine follicular fluid and serum reveals that excessive α(2)-macroglobulin in serum hampers successful expansion of cumulus-oocyte complexes. Proteomics 2009, 9, 4554–4565. [Google Scholar] [CrossRef] [PubMed]
- Fahiminiya, S.; Reynaud, K.; Labas, V.; Batard, S.; Chastant-Maillard, S.; Gerard, N. Steroid hormones content and proteomic analysis of canine follicular fluid during the preovulatory period. Reprod. Biol. Endocrinol. 2010, 8, 132. [Google Scholar] [CrossRef] [PubMed]
- Unwin, R.D.; Griffiths, J.R.; Whetton, A.D. Simultaneous analysis of relative protein expression levels across multiple samples using iTRAQ isobaric tags with 2D nano LC-MS/MS. Nat. Protoc. 2010, 5, 1574–1582. [Google Scholar] [CrossRef] [PubMed]
- Severino, V.; Malorni, L.; Cicatiello, A.E.; D'Esposito, V.; Longobardi, S.; Colacurci, N.; Miraglia, N.; Sannolo, N.; Farina, A.; Chambery, A. An integrated approach based on multiplexed protein array and iTRAQ labeling for in-depth identification of pathways associated to IVF outcome. PLoS ONE 2013, 8, e77303. [Google Scholar] [CrossRef] [PubMed]
- Zamah, A.M.; Hassis, M.E.; Albertolle, M.E.; Williams, K.E. Proteomic analysis of human follicular fluid from fertile women. Clin. Proteom. 2015, 12, 5. [Google Scholar] [CrossRef] [PubMed]
- Bijttebier, J.; Tilleman, K.; Deforce, D.; Dhaenens, M.; Van Soom, A.; Maes, D. Proteomic study to identify factors in follicular fluid and/or serum involved in in vitro cumulus expansion of porcine oocytes. Soc. Reprod. Fertil. Suppl. 2009, 66, 205–206. [Google Scholar] [PubMed]
- Vizcaino, J.A.; Csordas, A.; Del-Toro, N.; Dianes, J.A.; Griss, J.; Lavidas, I.; Mayer, G.; Perez-Riverol, Y.; Reisinger, F.; Ternent, T.; et al. 2016 update of the pride database and its related tools. Nucleic Acids Res. 2016, 44, D447–D456. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.L.; Anderson, N.G. The human plasma proteome: History, character, and diagnostic prospects. Mol. Cell. Proteom. 2002, 1, 845–867. [Google Scholar] [CrossRef]
- Huang, Y.L.; Huang, D.L.; Guan, J.L.; Pan, Q.; Fu, Q.; Huang, F. L.; Zhang, M.; Lu, K.H. Establishment of two dimensional electrophoresis method and mass spectrumetry analysis of the differential proteins of buffalo follicular fluid. Acta Vet. Zootech. Sin. 2013, 44, 1244–1250. [Google Scholar]
- Nesvizhskii, A.I.; Aebersold, R. Interpretation of shotgun proteomic data: The protein inference problem. Mol. Cell. Proteom. 2005, 4, 1419–1440. [Google Scholar] [CrossRef] [PubMed]
- Hanrieder, J.; Zuberovic, A.; Bergquist, J. Surface modified capillary electrophoresis combined with in solution isoelectric focusing and MALDI-TOF/TOF MS: A gel-free multidimensional electrophoresis approach for proteomic profiling—exemplified on human follicular fluid. J. Chromatogr. A 2009, 1216, 3621–3628. [Google Scholar] [CrossRef] [PubMed]
- Markstrom, E.; Svensson, E.; Shao, R.; Svanberg, B.; Billig, H. Survival factors regulating ovarian apoptosis—Dependence on follicle differentiation. Reproduction 2002, 123, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Aebersold, R. High-accuracy proteome maps of human body fluids. Genome Biol. 2006, 7, 242. [Google Scholar] [CrossRef] [PubMed]
- Schweigert, F.J.; Gericke, B.; Wolfram, W.; Kaisers, U.; Dudenhausen, J.W. Peptide and protein profiles in serum and follicular fluid of women undergoing ivf. Hum. Reprod. 2006, 21, 2960–2968. [Google Scholar] [CrossRef] [PubMed]
- Farrah, T.; Deutsch, E.W.; Omenn, G.S.; Campbell, D.S.; Sun, Z.; Bletz, J.A.; Mallick, P.; Katz, J.E.; Malmstrom, J.; Ossola, R.; et al. A high-confidence human plasma proteome reference set with estimated concentrations in peptideatlas. Mol. Cell. Proteom. 2011, 10. [Google Scholar] [CrossRef] [PubMed]
- Jarkovska, K.; Martinkova, J.; Liskova, L.; Halada, P.; Moos, J.; Rezabek, K.; Gadher, S.J.; Kovarova, H. Proteome mining of human follicular fluid reveals a crucial role of complement cascade and key biological pathways in women undergoing in vitro fertilization. J. Proteome Res. 2010, 9, 1289–1301. [Google Scholar] [CrossRef] [PubMed]
- Ebisch, I.M.; Thomas, C.M.; Wetzels, A.M.; Willemsen, W.N.; Sweep, F.C.; Steegers-Theunissen, R.P. Review of the role of the plasminogen activator system and vascular endothelial growth factor in subfertility. Fertil. Steril. 2008, 90, 2340–2350. [Google Scholar] [CrossRef] [PubMed]
- Espey, L.L. Current status of the hypothesis that mammalian ovulation is comparable to an inflammatory reaction. Biol. Reprod. 1994, 50, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Curry, T.E., Jr.; Osteen, K.G. The matrix metalloproteinase system: Changes, regulation, and impact throughout the ovarian and uterine reproductive cycle. Endocr. Rev. 2003, 24, 428–465. [Google Scholar] [CrossRef] [PubMed]
- Crews, S.T. Control of cell lineage-specific development and transcription by bHLH-PAS proteins. Genes Dev. 1998, 12, 607–620. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, P.H.; Ratcliffe, P.J. Oxygen sensors and angiogenesis. Semin. Cell Dev. Biol. 2002, 13, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Alam, H.; Maizels, E.T.; Park, Y.; Ghaey, S.; Feiger, Z.J.; Chandel, N.S.; Hunzicker-Dunn, M. Follicle-stimulating hormone activation of hypoxia-inducible factor-1 by the phosphatidylinositol 3-kinase/akt/ras homolog enriched in brain (Rheb)/mammalian target of rapamycin (mTOR) pathway is necessary for induction of select protein markers of follicular differentiation. J. Biol. Chem. 2004, 279, 19431–19440. [Google Scholar] [PubMed]
- Irving-Rodgers, H.F.; Rodgers, R.J. Extracellular matrix of the developing ovarian follicle. Semin. Reprod. Med. 2006, 24, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Irving-Rodgers, H.F.; Rodgers, R.J. Extracellular matrix in ovarian follicular development and disease. Cell Tissue Res. 2005, 322, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Kidder, G.M.; Vanderhyden, B.C. Bidirectional communication between oocytes and follicle cells: Ensuring oocyte developmental competence. Can. J. Physiol. Pharmacol. 2010, 88, 399–413. [Google Scholar] [CrossRef] [PubMed]
- Conti, M.; Hsieh, M.; Zamah, A.M.; Oh, J.S. Novel signaling mechanisms in the ovary during oocyte maturation and ovulation. Mol. Cell. Endocrinol. 2012, 356, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.G.; Ushizawa, K.; Hosoe, M.; Takahashi, T. Differential genome-wide gene expression profiling of bovine largest and second-largest follicles: Identification of genes associated with growth of dominant follicles. Reprod. Biol. Endocrinol. 2010, 8, 11. [Google Scholar] [CrossRef] [PubMed]
- Stilley, J.A.; Sharpe-Timms, K.L. TIMP1 contributes to ovarian anomalies in both an MMP-dependent and -independent manner in a rat model. Biol. Reprod. 2012, 86, 47. [Google Scholar] [CrossRef] [PubMed]
- Kwintkiewicz, J.; Giudice, L.C. The interplay of insulin-like growth factors, gonadotropins, and endocrine disruptors in ovarian follicular development and function. Semin. Reprod. Med. 2009, 27, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, A.; Patel, O.V.; Lee, K.B.; Park, K.E.; Salem, M.; Yao, J.; Ireland, J.J.; Smith, G.W. Identification of novel bovine cumulus cell molecular markers predictive of oocyte competence: Functional and diagnostic implications. Biol. Reprod. 2008, 79, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Kawamura, K.; Deguchi, M.; Takae, S.; Mulders, S.M.; Hsueh, A.J. Intraovarian thrombin and activated protein C signaling system regulates steroidogenesis during the periovulatory period. Mol. Endocrinol. 2012, 26, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Hamel, M.; Dufort, I.; Robert, C.; Gravel, C.; Leveille, M.C.; Leader, A.; Sirard, M.A. Identification of differentially expressed markers in human follicular cells associated with competent oocytes. Hum. Reprod. 2008, 23, 1118–1127. [Google Scholar] [CrossRef] [PubMed]
- Matos, L.; Stevenson, D.; Gomes, F.; Silva-Carvalho, J.L.; Almeida, H. Superoxide dismutase expression in human cumulus oophorus cells. Mol. Hum. Reprod. 2009, 15, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Ryu, K.Y.; Sinnar, S.A.; Reinholdt, L.G.; Vaccari, S.; Hall, S.; Garcia, M.A.; Zaitseva, T.S.; Bouley, D.M.; Boekelheide, K.; Handel, M.A.; et al. The mouse polyubiquitin gene ubb is essential for meiotic progression. Mol. Cell. Biol. 2008, 28, 1136–1146. [Google Scholar] [CrossRef] [PubMed]
- Tsuiki, A.; Preyer, J.; Hung, T.T. Fibronectin and glycosaminoglycans in human preovulatory follicular fluid and their correlation to follicular maturation. Hum. Reprod. 1988, 3, 425–429. [Google Scholar] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. String v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Krebiehl, G.; Ruckerbauer, S.; Burbulla, L.F.; Kieper, N.; Maurer, B.; Waak, J.; Wolburg, H.; Gizatullina, Z.; Gellerich, F.N.; Woitalla, D.; et al. Reduced basal autophagy and impaired mitochondrial dynamics due to loss of parkinson’s disease-associated protein dj-1. PLoS ONE 2010, 5, e9367. [Google Scholar] [CrossRef] [PubMed]
- Argenzio, E.; Margadant, C.; Leyton-Puig, D.; Janssen, H.; Jalink, K.; Sonnenberg, A.; Moolenaar, W.H. Clic4 regulates cell adhesion and β1 integrin trafficking. J. Cell Sci. 2014, 127, 5189–5203. [Google Scholar] [CrossRef] [PubMed]
- Krisher, R.L. The effect of oocyte quality on development. J. Anim. Sci. 2004, 82, E14–E23. [Google Scholar] [PubMed]
- Sturmey, R.G.; Reis, A.; Leese, H.J.; McEvoy, T.G. Role of fatty acids in energy provision during oocyte maturation and early embryo development. Reprod. Domest. Anim. 2009, 44, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Paczkowski, M.; Krisher, R. Aberrant protein expression is associated with decreased developmental potential in porcine cumulus-oocyte complexes. Mol. Reprod. Dev. 2010, 77, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, E.; Weber, K. Intermediate filaments: Structure, dynamics, function, and disease. Annu. Rev. Biochem. 1994, 63, 345–382. [Google Scholar] [CrossRef] [PubMed]
- Sickles, D.W.; Sperry, A.O.; Testino, A.; Friedman, M. Acrylamide effects on kinesin-related proteins of the mitotic/meiotic spindle. Toxicol. Appl. Pharmacol. 2007, 222, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.J.; Patel, S.; Eriksson, J.E.; Kraemer, F.B. Vimentin is a functional partner of hormone sensitive lipase and facilitates lipolysis. J. Proteome Res. 2010, 9, 1786–1794. [Google Scholar] [CrossRef] [PubMed]
- Iwatsuki, H.; Suda, M. Seven kinds of intermediate filament networks in the cytoplasm of polarized cells: Structure and function. Acta Histochem. Cytochem. 2010, 43, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Marceau, N.; Schutte, B.; Gilbert, S.; Loranger, A.; Henfling, M.E.; Broers, J.L.; Mathew, J.; Ramaekers, F.C. Dual roles of intermediate filaments in apoptosis. Exp. Cell Res. 2007, 313, 2265–2281. [Google Scholar] [CrossRef] [PubMed]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. Kobas 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.L.; Fu, Q.; Yang, L.; Guan, J.L.; Pan, H.; Chen, F.M.; Lu, K.L.; Zhang, M. Differences between high- and low-motility buffalo sperm identified by comparative proteomics. Reprod. Domest. Anim. 2015, 50, 443–451. [Google Scholar] [CrossRef] [PubMed]










| Large Follicles | Small Follicles | |
|---|---|---|
| Diameter, mm | >8 | <4 |
| Numbers, n | n = 7 | n = 25 |
| Estradiol, ng/mL | 131.41 ± 54.70 | 24.64 ± 6.35 |
| Progesterone, ng/mL | 33.35 ± 7.55 | 13.08 ± 3.28 |
| E2/P4 | 3.93 ± 1.72 | 1.89 ± 0.26 |
| KEGG ID | Pathway Name | Protein Counts | p-Value * |
|---|---|---|---|
| mmu04610 | Complement and coagulation cascades | 33 | 8.63 × 10−27 |
| mmu03010 | Ribosome | 21 | 8.31 × 10−10 |
| mmu01200 | Carbon metabolism | 13 | 1.08 × 10−5 |
| mmu00010 | Glycolysis/Gluconeogenesis | 10 | 4.05 × 10−5 |
| mmu00030 | Pentose phosphate pathway | 6 | 0.00015 |
| mmu01230 | Biosynthesis of amino acids | 9 | 0.00028 |
| mmu03050 | Proteasome | 7 | 0.00040 |
| mmu00051 | Fructose and mannose metabolism | 6 | 0.00176 |
| mmu03040 | Spliceosome | 10 | 0.00319 |
| mmu00480 | Glutathione metabolism | 6 | 0.00327 |
| mmu00020 | Citrate cycle (TCA cycle) | 4 | 0.00699 |
| mmu04612 | Antigen processing and presentation | 7 | 0.01812 |
| mmu04512 | ECM-receptor interaction | 7 | 0.02425 |
| mmu04810 | Regulation of actin cytoskeleton | 13 | 0.02786 |
| mmu04145 | Phagosome | 10 | 0.03001 |
| mmu04066 | HIF-1 signaling pathway | 8 | 0.04746 |
| Accession | Protein Name | Gene Symbol | Fold Change * | No. of Unique Peptides | Sequence Coverage % | Functions |
|---|---|---|---|---|---|---|
| Q2KJD0 | Tubulin β-5 chain | TUBB5 | 13.69/3.08 Δ | 4 | 11.71 | The major constituent of microtubules |
| A6QLL8 | Fructose-bisphosphate aldolase | ALDOA | 8.22/3.06 Δ | 3 | 8.79 | Involved in glycolysis and energy metabolism |
| Q3SZR3 | α-1-acid glycoprotein | ORM1 | 3.06/2.32 Δ | 11 | 2 | Functions as a transport protein in the blood |
| E1BH06 | complement C4 | C4A | 0.44/0.36 ∇ | 10 | 17.7 | Non-enzymatic component of C3 and C5 convertases and thus essential for propagation of the classical complement pathway |
| F1MSZ6 | Antithrombin-III | SERPINC1 | 0.44/0.40 ∇ | 13 | 26.9 | Most important serine protease inhibitor in plasma that regulates the blood coagulation cascade |
| Q7SIH1 | α-2-macroglobulin | A2M | 0.31/0.50 ∇ | 19 | 14.1 | Can inhibit all four classes of proteinases |
| Q28085 | Complement factor H | CFH | 0.35/0.48 ∇ | 4 | 9.55 | Functions as a cofactor in the inactivation of C3b by factor I in the alternative complement pathway |
| P48616 | Vimentin | VIM | 7.78/3.89 Δ | 4 | 7.08 | Class III intermediate filament attached to the nucleus, endoplasmic reticulum, and mitochondria |
| G3N0V0 | Uncharacterized protein (fragment) | unknown | 4.60/6.29 Δ | 7 | 20.55 | Uncharacterized protein |
| A6QPP2 | SERPIND1 protein | SERPIND1 | 0.29/0.44 ∇ | 6 | 13.1 | Heparin cofactor 2 precursor |
| Q5E947 | Peroxiredoxin-1 | PRDX1 | 3.66/3.93 Δ | 2 | 9.55 | Involved in redox regulation of the cell. Might play an important role in eliminating peroxides generated during metabolism |
| Uniprot Accession No. | Protein Name | Protein Symbol |
|---|---|---|
| Q05717 | Insulin-like growth factor-binding protein 5 | IGFBP5 |
| F1MUK3 | Insulin-like growth factor-binding protein 6 | IGFBP6 |
| F1MPP2 | Insulin-like growth factor-binding protein 7 | IGFBP7 |
| F1MJZ4 | Insulin-like growth factor-binding protein complex acid labile subunit | IGFals |
| F1N430 | Metalloproteinase inhibitor 2 | TIMP2 |
| P62894 | Cytochrome c | CYCS |
| Q2KIF2 | Leucine-rich α-2-glycoprotein 1 | LRG1 |
| F1MD95 | Calsyntenin-1 | CLSTN1 |
| A1A4K5 | Ectonucleotidepyrophosphatase/phosphodiesterase family member 2 | ENPP2 |
| P13696 | Phosphatidylethanolamine-binding protein 1 | PEBP1 |
| Q95121 | Pigment epithelium-derived factor | SERPINF1 |
| Protein Name | Protein Symbol | Reported Association of Biomarker |
|---|---|---|
| Fibronectin (P07589) | FN1 | Oocytes maturation [53] |
| Uncharacterized protein (F1MZX2) | SERPINE2 | Positively associated with pregnancy outcome [50] |
| Superoxide dismutase (A3KLR9) | SOD | Correlated with success of assisted reproductive techniques [51] |
| Cathepsin B (P07688) | CTSB | Poor quality in cattle oocyte [48] |
| Polyubiquitin-B (P0CG53) | UBB | Abnormal oocyte morphology; decreased oocyte number; thin zonapellucida [52] |
| Thrombin (P00735) | F2 | Optimal follicular luteinization in mice [49] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, Q.; Huang, Y.; Wang, Z.; Chen, F.; Huang, D.; Lu, Y.; Liang, X.; Zhang, M. Proteome Profile and Quantitative Proteomic Analysis of Buffalo (Bubalusbubalis) Follicular Fluid during Follicle Development. Int. J. Mol. Sci. 2016, 17, 618. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17050618
Fu Q, Huang Y, Wang Z, Chen F, Huang D, Lu Y, Liang X, Zhang M. Proteome Profile and Quantitative Proteomic Analysis of Buffalo (Bubalusbubalis) Follicular Fluid during Follicle Development. International Journal of Molecular Sciences. 2016; 17(5):618. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17050618
Chicago/Turabian StyleFu, Qiang, Yulin Huang, Zhiqiang Wang, Fumei Chen, Delun Huang, Yangqing Lu, Xianwei Liang, and Ming Zhang. 2016. "Proteome Profile and Quantitative Proteomic Analysis of Buffalo (Bubalusbubalis) Follicular Fluid during Follicle Development" International Journal of Molecular Sciences 17, no. 5: 618. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17050618