Expression Patterns and Functional Novelty of Ribonuclease 1 in Herbivorous Megalobrama amblycephala
Abstract
:1. Introduction
2. Results
2.1. Protein Alignment
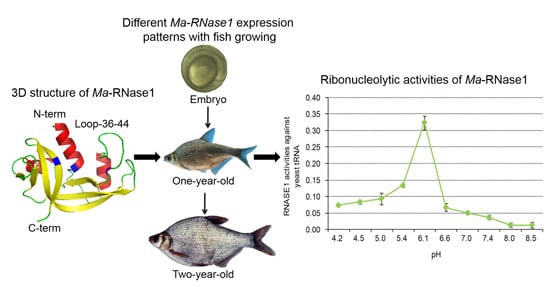
2.2. Structural Predictions
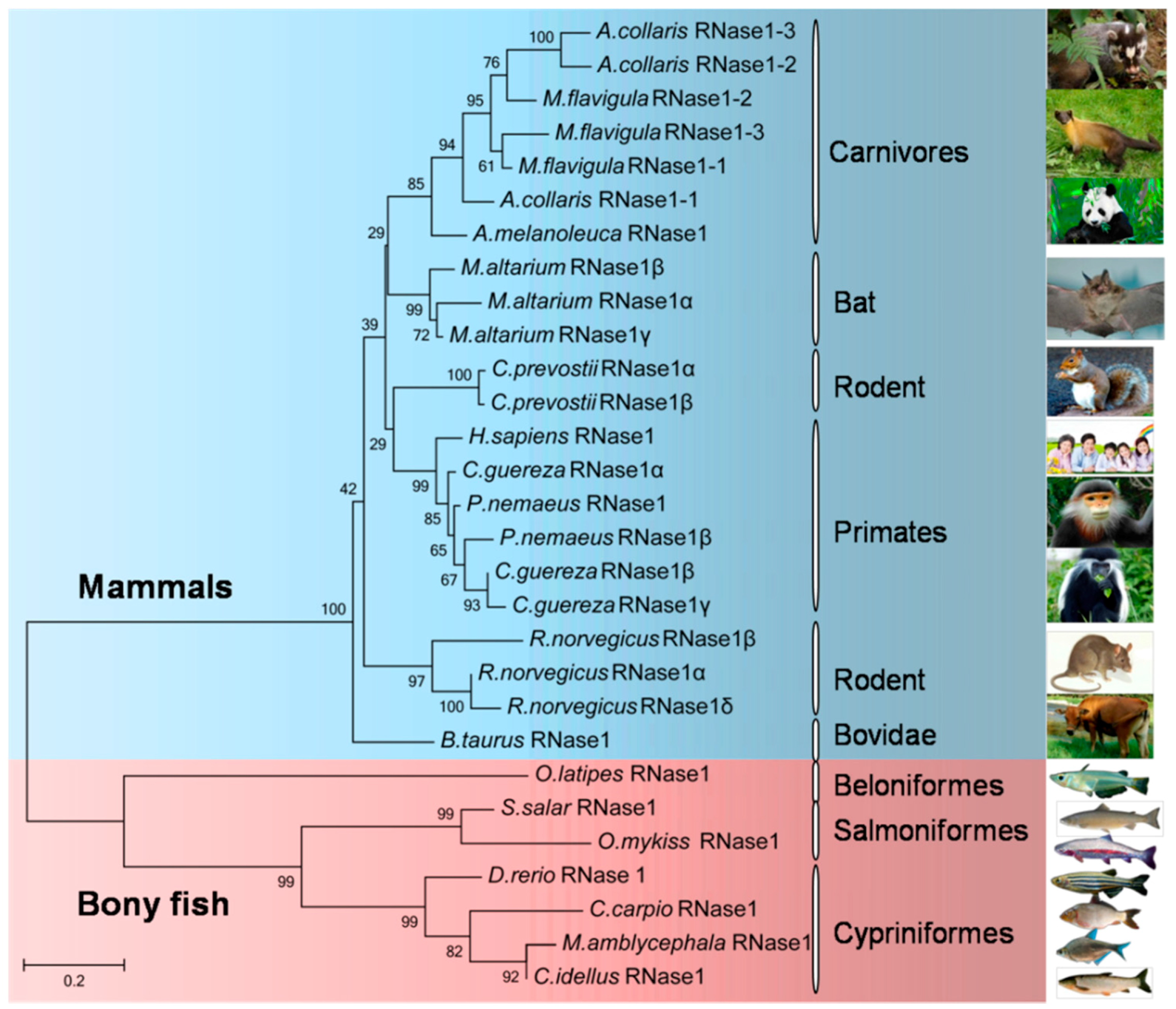
2.3. Phylogenetic Relationships among the Bony Fish and Mammalian RNase1 Proteins
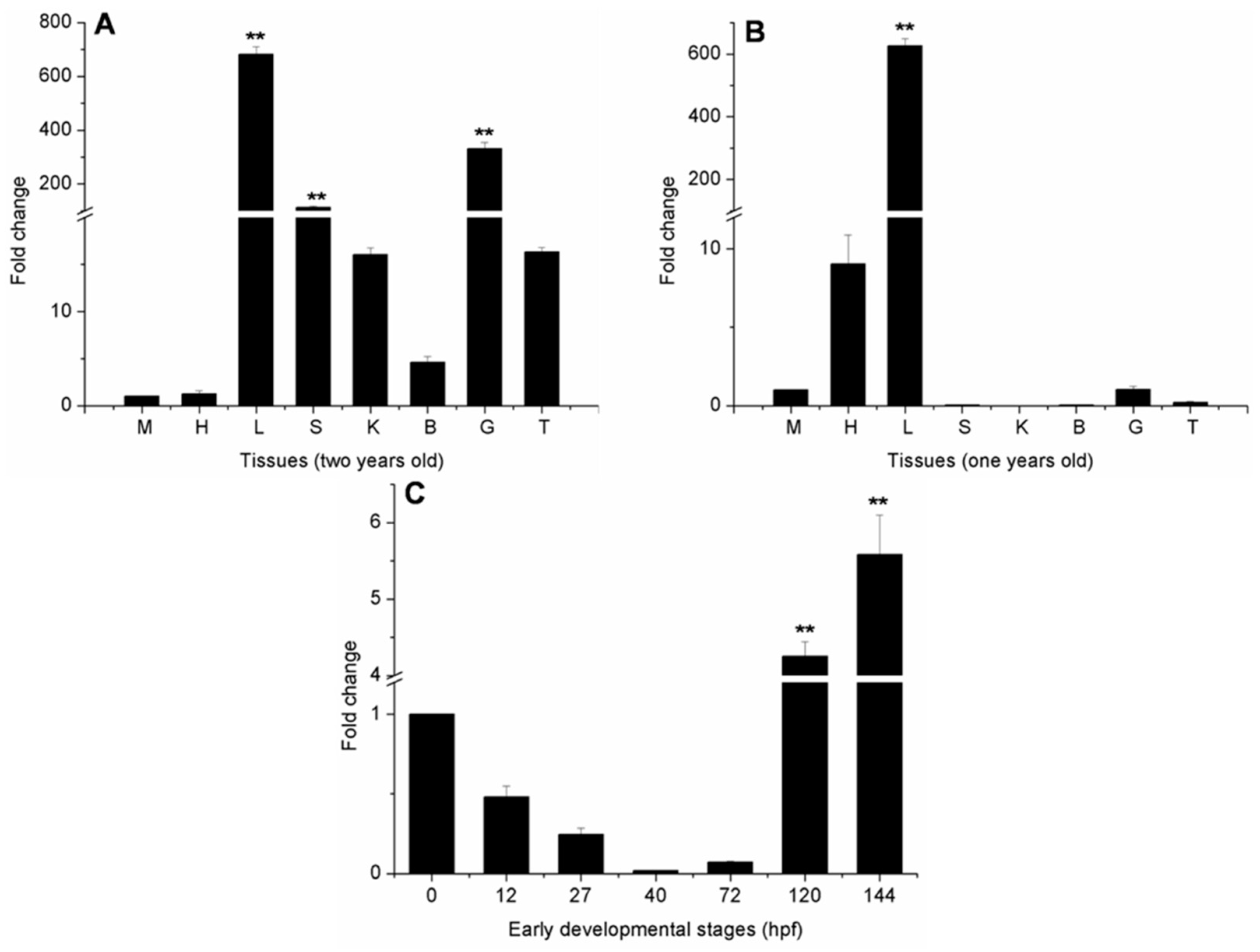
2.4. Ma-RNase1 Expression Patterns
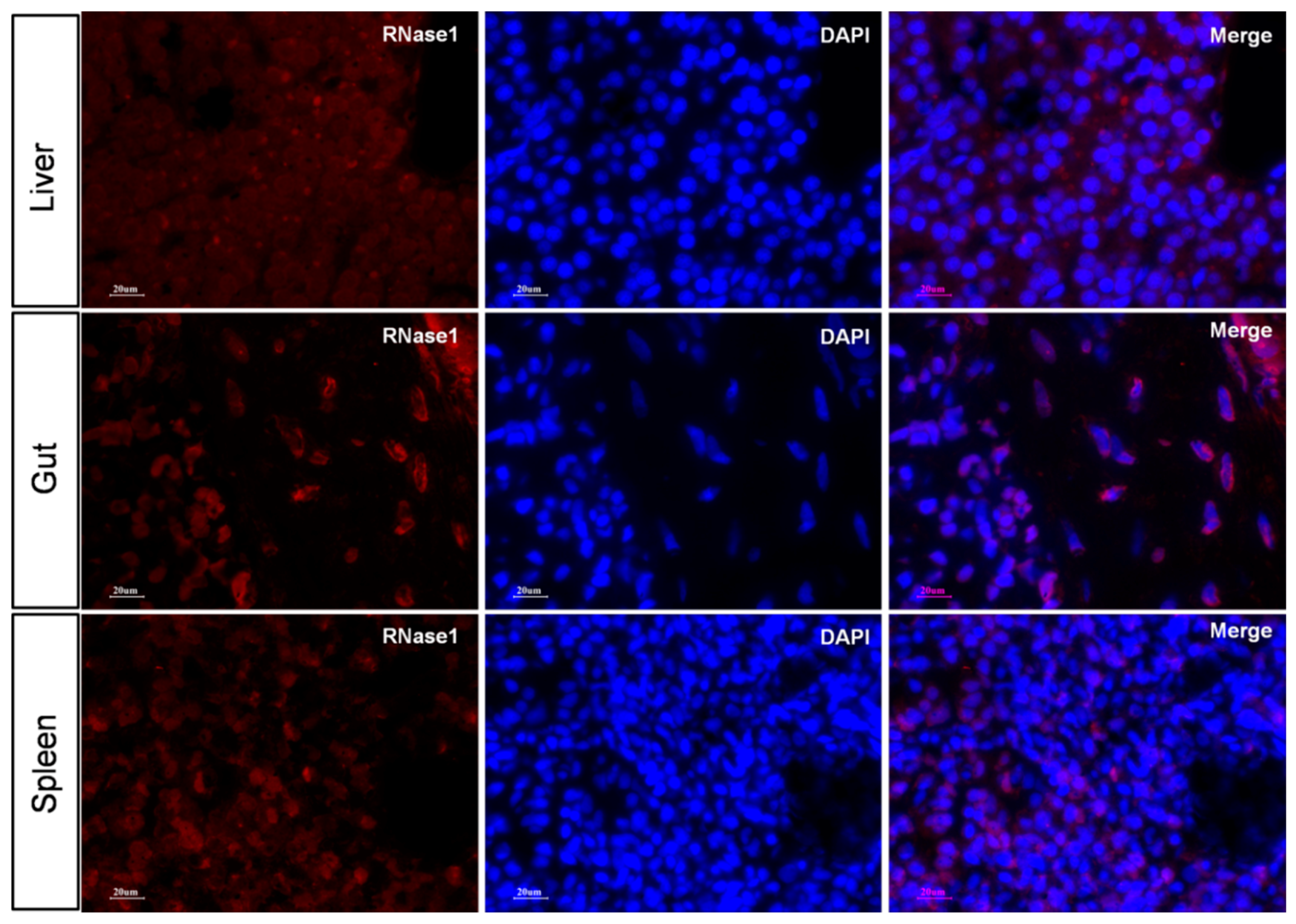
2.5. Ma-RNase1 Peptide Production Analyzed by Immunohistochemistry
2.6. Ribonucleolytic Activity of Ma-RNase1
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Identification of RNase1 Genes
4.3. Protein Alignment
4.4. Structural Predictions
4.5. Phylogenetic Analyses
4.6. Ma-RNase1 Gene Expression Analysis
4.7. Assessment of Ma-RNase1 Production by Immunostaining
4.8. Recombinant Protein Preparation
4.9. Assays of Ma-RNase1 Activity
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sorrentino, S. The eight human “canonical” ribonucleases: Molecular diversity, catalytic properties, and special biological actions of the enzyme proteins. FEBS Lett. 2010, 584, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, H.F. RNase A ribonucleases and host defense: An evolving story. J. Leukoc. Biol. 2008, 83, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Barnard, E.A. Biological function of pancreatic ribonuclease. Nature 1969, 221, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, Y.; Rosenberg, H.F. Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat. Genet. 2002, 30, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Dyer, K.D.; Rosenberg, H.F. The RNase a superfamily: Generation of diversity and innate host defense. Mol. Divers. 2006, 10, 585–597. [Google Scholar] [CrossRef] [PubMed]
- Monti, D.M.; Yu, W.; Pizzo, E.; Shima, K.; Hu, M.G.; di Malta, C.; Piccoli, R.; D’Alessio, G.; Hu, G.F. Characterization of the angiogenic activity of zebrafish ribonucleases. FEBS J. 2009, 276, 4077–4090. [Google Scholar] [CrossRef] [PubMed]
- Pizzo, E.; Merlino, A.; Turano, M.; Krauss, R.I.; Coscia, F.; Zanfardino, A.; Varcamonti, M.; Furia, A.; Giancola, C.; Mazzarella, L.; et al. A new RNase sheds light on the RNase/angiogenin subfamily from zebrafish. Biochem. J. 2011, 433, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Pizzo, E.; Varcamonti, M.; Maro, A.D.; Zanfardino, A.; Giancola, C.; D’Alessio, G. Ribonucleases with angiogenic and bactericidal activities from the Atlantic salmon. FEBS J. 2008, 275, 1283–1295. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Badia, M.; Moussaoui, M.; Sanchez, D.; Nogués, M.; Boix, E. Comparison of human RNase 3 and RNase 7 bactericidal action at the Gram-negative and Gram-positive bacterial cell wall. FEBS J. 2010, 277, 1713–1725. [Google Scholar] [CrossRef] [PubMed]
- Futami, J.; Tsushima, Y.; Murato, Y.; Tada, H.; Sasaki, J.; Seno, M.; Yamada, H. Tissue-specific expression of pancreatic-type RNases and RNase inhibitor in humans. DNA Cell Biol. 1997, 16, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Beintema, J.J. The primary structure of langur (Presbytis entellus) pancreatic ribonuclease: Adaptive features in digestive enzymes in mammals. Mol. Biol. Evol. 1990, 7, 470–477. [Google Scholar] [PubMed]
- Sorrentino, S.; Naddeo, M.; Russo, A.; D’Alessio, G. Degradation of double-stranded RNA by human pancreatic ribonuclease: Crucial role of noncatalytic basic amino acid residues. Biochemistry 2003, 42, 10182–10190. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. Parallel adaptive origins of digestive RNases in Asian and African leaf monkeys. Nat. Genet. 2006, 38, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Dubois, J.Y.F.; Jekel, P.A.; Mulder, P.P.; Bussink, A.P.; Catzeflis, F.M.; Carsana, A.; Beintema, J.J. Pancreatic-type ribonuclease 1 gene duplications in rat species. J. Mol. Evol. 2002, 55, 522–533. [Google Scholar] [CrossRef] [PubMed]
- Siegel, S.J.; Percopo, C.M.; Dyer, K.D.; Zhao, W.; Roth, V.L.; Mercer, J.M.; Rosenberg, H.F. RNase 1 genes from the family Sciuridae define a novel rodent ribonuclease cluster. Mamm. Genome 2009, 20, 749–757. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Liu, Y.; Meng, F.; He, B.; Han, N.; Li, G.; Rossiter, S.J.; Zhang, S. Multiple bursts of pancreatic ribonuclease gene duplication in insect-eating bats. Gene 2013, 526, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Zhang, Y.P. The unusual adaptive expansion of pancreatic ribonuclease gene in carnivora. Mol. Biol. Evol. 2006, 23, 2326–2335. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, X.P.; Cho, S.; Lim, B.K.; Irwin, D.M.; Ryder, O.A.; Zhang, Y.P.; Yu, L. Evolutionary and functional novelty of pancreatic ribonuclease: A study of musteloidea (order Carnivora). Sci. Rep. 2014, 4, 5074. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Zhang, J. Zebrafish ribonucleases are bactericidal: Implications for the origin of the vertebrate RNase a superfamily. Mol. Biol. Evol. 2007, 24, 1259–1268. [Google Scholar] [CrossRef] [PubMed]
- Pizzo, E.; Buonanno, P.; di Maro, A.; Ponticelli, S.; de Falco, S.; Quarto, N.; Cubellis, M.V.; D’Alessio, G. Ribonucleases and angiogenins from fish. J. Biol. Chem. 2006, 281, 27454–27460. [Google Scholar] [CrossRef] [PubMed]
- Kazakou, K.; Holloway, D.E.; Prior, S.H.; Subramanian, V.; Acharya, K.R. Ribonuclease A homologues of the zebrafish: Polymorphism, crystal structures of two representatives and their evolutionary implications. J. Mol. Biol. 2008, 380, 206–222. [Google Scholar] [CrossRef] [PubMed]
- Kövér, K.E.; Bruix, M.; Santoro, J.; Batta, G.; Laurents, D.V.; Rico, M. The solution structure and dynamics of human pancreatic ribonuclease determined by NMR spectroscopy provide insight into its remarkable biological activities and inhibition. J. Mol. Biol. 2008, 379, 953–965. [Google Scholar] [CrossRef] [PubMed]
- Strydom, D.J. The angiogenins. Cell. Mol. Life Sci. 1998, 54, 811–824. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Beintema, J.J.; Zhang, J. The ribonuclease a superfamily of mammals and birds: Identifying new members and tracing evolutionary histories. Genomics 2005, 85, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Dubois, J.Y.F.; Ursing, B.M.; Kolkman, J.A.; Beintema, J.J. Molecular evolution of mammalian ribonucleases 1. Mol. Phylogenet. Evol. 2003, 27, 453–463. [Google Scholar] [CrossRef]
- Rosenberg, H.F.; Dyer, K.D. Eosinophil cationic protein and eosinophil-derived neurotoxin: Evolution of novel function in a primate ribonuclease gene family. J. Biol. Chem. 1995, 270, 21539–21544. [Google Scholar] [CrossRef] [PubMed]
- Nitto, T.; Dyer, K.D.; Czapiga, M.; Rosenberg, H.F. Evolution and function of leukocyte RNase A ribonucleases of the avian species, Gallus gallus. J. Biol. Chem. 2006, 281, 25622–25634. [Google Scholar] [CrossRef] [PubMed]
- Kent, W.J. BLAT—The BLAST-like alignment tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Birney, E.; Clamp, M.; Durbin, R. GeneWise and Genomewise. Genome Res. 2004, 14, 988–995. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- SignalP 4.1 Server. Available online: http://www.cbs.dtu.dk/services/SignalP/ (accessed on 20 March 2015).
- Compute pI/Mw Tool. Available online: http://web.expasy.org/compute_pi/ (accessed on 20 March 2015).
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System; DeLano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]




© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Wang, W. Expression Patterns and Functional Novelty of Ribonuclease 1 in Herbivorous Megalobrama amblycephala. Int. J. Mol. Sci. 2016, 17, 786. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17050786
Liu H, Wang W. Expression Patterns and Functional Novelty of Ribonuclease 1 in Herbivorous Megalobrama amblycephala. International Journal of Molecular Sciences. 2016; 17(5):786. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17050786
Chicago/Turabian StyleLiu, Han, and Weimin Wang. 2016. "Expression Patterns and Functional Novelty of Ribonuclease 1 in Herbivorous Megalobrama amblycephala" International Journal of Molecular Sciences 17, no. 5: 786. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17050786