Metabolomic Studies of Oral Biofilm, Oral Cancer, and Beyond
Abstract
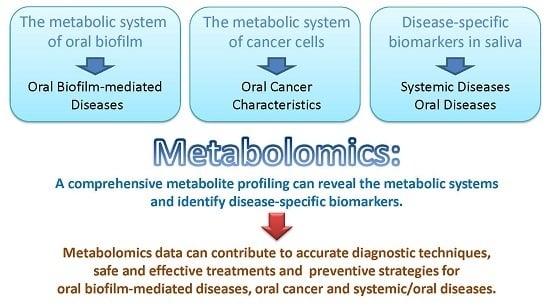
:1. Introduction
1.1. What Is Metabolomics?
1.2. Advantages of CE-TOFMS-Based Metabolomics for Oral Biofilm and Oral Cancer Research
2. Metabolomics Research into the Oral Biofilm
2.1. Sample Preparation for Metabolomic Analysis and the Study Method
2.2. Metabolomics of the Oral Biofilm and a Comparison with the Findings for Representative Oral Bacteria
2.3. Effects of Fluoride and Xylitol on Metabolism in the Oral Biofilm
3. Application to Oral Cancer Research
4. Application to Saliva Samples
5. Conclusions and the Future Research
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Soga, T.; Ueno, Y.; Naraoka, H.; Ohashi, Y.; Tomita, M.; Nishioka, T. Simultaneous determination of anionic intermediates for Bacillus subtilis metabolic pathways by capillary electrophoresis electrospray ionization mass spectrometry. Anal. Chem. 2002, 74, 2233–2239. [Google Scholar] [CrossRef] [PubMed]
- Soga, T.; Ohashi, Y.; Ueno, Y.; Naraoka, H.; Tomita, M.; Nishioka, T. Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. J. Proteome Res. 2003, 2, 488–494. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, Y.; Hirayama, A.; Ishikawa, T.; Nakamura, S.; Shimizu, K.; Ueno, Y.; Tomita, M.; Soga, T. Depiction of metabolome changes in histidine-starved Escherichia coli by CE-TOFMS. Mol. Biosyst. 2008, 4, 35–147. [Google Scholar] [CrossRef] [PubMed]
- Morohashi, M.; Ohashi, Y.; Tani, S.; Ishii, K.; Itaya, M.; Nanamiya, H.; Kawamura, F.; Tomita, M.; Soga, T. Model based definition of population heterogeneity and its effects on metabolism in sporulating Bacillus subtilis. J. Biochem. 2007, 142, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, A.; Kami, K.; Sugimoto, M.; Sugawara, M.; Toki, N.; Onozuka, H.; Kinoshita, T.; Saito, N.; Ochiai, A.; Tomita, M.; et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009, 69, 4918–4925. [Google Scholar] [CrossRef] [PubMed]
- Minami, Y.; Kasukawa, T.; Kakazu, Y.; Iigo, M.; Sugimoto, M.; Ikeda, S.; Yasui, A.; van der Horst, G.T.; Soga, T.; Ueda, H.R. Measurement of internal body time by blood metabolomics. Proc. Natl. Acad. Sci. USA 2009, 106, 9890–9895. [Google Scholar] [CrossRef] [PubMed]
- Soga, T.; Baran, R.; Suematsu, M.; Ueno, Y.; Ikeda, S.; Sakurakawa, T.; Kakazu, Y.; Ishikawa, T.; Robert, M.; Nishioka, T.; et al. Differential metabolomics reveals ophthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption. J. Biol. Chem. 2006, 281, 16768–16776. [Google Scholar] [CrossRef] [PubMed]
- Marsh, P.D. Microbial ecology of dental plaque and its significance in health and disease. Adv. Dent. Res. 1994, 8, 263–271. [Google Scholar] [PubMed]
- Tanzer, J.M.; Livingston, J.; Thompson, A.M. The microbiology of primary dental caries in humans. J. Dent. Educ. 2001, 65, 1028–1037. [Google Scholar] [PubMed]
- Becker, M.R.; Paster, B.J.; Leys, E.J.; Moeschberger, M.L.; Kenyon, S.G.; Galvin, J.L.; Boches, S.K.; Dewhirst, F.E.; Griffen, A.L. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 2002, 40, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Chhour, K.L.; Nadkarni, M.A.; Byun, R.; Martin, F.E.; Jacques, N.A.; Hunter, N. Molecular analysis of microbial diversity in advanced caries. J. Clin. Microbiol. 2005, 43, 843–849. [Google Scholar] [CrossRef] [PubMed]
- Ledder, R.G.; Gilbert, P.; Huws, S.A.; Aarons, L.; Ashley, M.P.; Hull, P.S.; McBain, A.J. Molecular analysis of the subgingival microbiota in health and disease. Appl. Environ. Microbiol. 2007, 73, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N. Oral microbiome metabolism: From “who are they?” to “what are they doing?”. J. Dent. Res. 2015, 94, 1628–1637. [Google Scholar] [CrossRef] [PubMed]
- Keyes, P.H. Research in dental caries. J. Am. Dent. Assoc. 1968, 76, 1357–1373. [Google Scholar] [CrossRef] [PubMed]
- Helgeland, K. Inhibitory effect of NH4Cl on secretion of collagen in human gingival fibroblasts. Scand. J. Dent. Res. 1984, 92, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Kurita-Ochiai, T.; Seto, S.; Suzuki, N.; Yamamoto, M.; Otsuka, K.; Abe, K.; Ochiai, K. Butyric acid induces apoptosis in inflamed fibroblasts. J. Dent. Res. 2008, 87, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Yaegaki, K.; Qian, W.; Murata, T.; Imai, T.; Sato, T.; Tanaka, T.; Kamoda, T. Oral malodorous compound causes apoptosis and genomic DNA damage in human gingival fibroblasts. J. Periodontal Res. 2008, 43, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Minakami, S.; Suzuki, C.; Saito, T.; Yoshikawa, H. Studies on erythrocyte glycolysis. I. Determination of the glycolytic intermediates in human erythrocytes. J. Biochem. 1965, 58, 543–550. [Google Scholar] [PubMed]
- Iwami, Y.; Yamada, T.; Araya, S. Glycolytic intermediates in Streptococcus mutans PK 1. Arch. Oral Biol. 1975, 20, 695–697. [Google Scholar] [CrossRef]
- Yamada, T.; Carlsson, J. Regulation of lactate dehydrogenase and change of fermentation products in streptococci. J. Bacteriol. 1975, 124, 55–61. [Google Scholar] [PubMed]
- Yamada, T.; Carlsson, J. Glucose-6-phosphate-dependent pyruvate kinase in Streptococcus mutans. J. Bacteriol. 1975, 124, 562–563. [Google Scholar] [PubMed]
- Iwami, Y.; Yamada, T. Rate-limiting steps of the glycolytic pathway in the oral bacteria Streptococcus mutans and Streptococcus sanguis and the influence of acidic pH on the glucose metabolism. Arch. Oral Biol. 1980, 25, 163–169. [Google Scholar] [CrossRef]
- Abbe, K.; Takahashi, S.; Yamada, T. Involvement of oxygen-sensitive pyruvate formate-lyase in mixed-acid fermentation by Streptococcus mutans under strictly anaerobic conditions. J. Bacteriol. 1982, 152, 175–182. [Google Scholar] [PubMed]
- Abbe, K.; Takahashi, S.; Yamada, T. Purification and properties of pyruvate kinase from Streptococcus sanguis and activator specificity of pyruvate kinase from oral streptococci. Infect. Immun. 1983, 39, 1007–1014. [Google Scholar] [PubMed]
- Iwami, Y.; Yamada, T. Regulation of glycolytic rate in Streptococcus sanguis grown under glucose-limited and glucose-excess conditions in a chemostat. Infect. Immun. 1985, 50, 378–381. [Google Scholar] [PubMed]
- Takahashi, N.; Iwami, Y.; Yamada, T. Metabolism of intracellular polysaccharide in the cells of Streptococcus mutans under strictly anaerobic conditions. Oral Microbiol. Immunol. 1991, 6, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Yamada, T. Stimulatory effect of bicarbonate on the glycolysis of Actinomyces viscosus and its biochemical mechanism. Oral Microbiol. Immunol. 1992, 7, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Iwami, Y.; Abbe, K.; Takahashi-Abbe, S.; Yamada, T. Acid production by streptococci growing at low pH in a chemostat under anaerobic conditions. Oral Microbiol. Immunol. 1992, 7, 304–308. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Yamada, T. Glucose and lactate metabolism by Actinomyces naeslundii. Crit. Rev. Oral Biol. Med. 1999, 10, 487–503. [Google Scholar] [CrossRef] [PubMed]
- Iwami, Y.; Takahashi-Abbe, S.; Takahashi, N.; Abbe, K.; Yamada, T. Rate-limiting steps of glucose and sorbitol metabolism in Streptococcus mutans cells exposed to air. Oral Microbiol. Immunol. 2000, 15, 325–328. [Google Scholar] [CrossRef] [PubMed]
- Hata, S.; Iwami, Y.; Kamiyama, K.; Yamada, T. Biochemical mechanisms of enhanced inhibition of fluoride on the anaerobic sugar metabolism by Streptococcus sanguis. J. Dent. Res. 1990, 69, 1244–1247. [Google Scholar] [CrossRef] [PubMed]
- Miyasawa, H.; Iwami, Y.; Mayanagi, H.; Takahashi, N. Xylitol inhibition of anaerobic acid production by Streptococcus mutans at various pH levels. Oral Microbiol. Immunol. 2003, 18, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Kakuta, H.; Iwami, Y.; Mayanagi, H.; Takahashi, N. Xylitol inhibition of acid production and growth of mutans streptococci in the presence of various dietary sugars under strictly anaerobic conditions. Caries Res. 2003, 37, 404–409. [Google Scholar] [CrossRef] [PubMed]
- Maehara, H.; Iwami, Y.; Mayanagi, H.; Takahashi, N. Synergistic inhibition by combination of fluoride and xylitol on glycolysis by mutans streptococci and its biochemical mechanism. Caries Res. 2005, 39, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Miyasawa-Hori, H.; Aizawa, S.; Takahashi, N. Difference in the xylitol sensitivity of acid production among Streptococcus mutans strains and the biochemical mechanism. Oral Microbiol. Immunol. 2006, 21, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Sato, T.; Yamada, T. Metabolic pathways for cytotoxic end product formation from glutamate—and aspartate-containing peptides by Porphyromonas gingivalis. J. Bacteriol. 2000, 182, 4704–4710. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Keenan, M.M.; Chi, J.T. Alternative fuels for cancer cells. Cancer J. 2015, 21, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Washio, J.; Mayanagi, G. Metabolomics of supragingival plaque and oral bacteria. J. Dent. Res. 2010, 89, 1383–1388. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Washio, J. Metabolomic effects of xylitol and fluoride on plaque biofilm in vivo. J. Dent. Res. 2011, 90, 1463–1468. [Google Scholar] [CrossRef] [PubMed]
- Kolenbrander, P.E. Oral microbial communities: Biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 2000, 54, 413–437. [Google Scholar] [CrossRef] [PubMed]
- Ximénez-Fyvie, L.A.; Haffajee, A.D.; Socransky, S.S. Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J. Clin. Periodontol. 2000, 27, 648–657. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, I.R. Biochemical effects of fluoride on oral bacteria. J. Dent. Res. 1990, 69, 660–667. [Google Scholar] [CrossRef] [PubMed]
- Marquis, R.E. Diminished acid tolerance of plaque bacteria caused by fluoride. J. Dent. Res. 1990, 69, 672–675. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, G.N. Review of fluoride research since 1959. Arch. Oral Biol. 1999, 44, 985–992. [Google Scholar] [CrossRef]
- Tatevossian, A. Fluoride in dental plaque and its effects. J. Dent. Res. 1990, 69, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Vogel, G.L.; Zhang, Z.; Chow, L.C.; Schumacher, G.E. Changes in lactate and other ions in plaque and saliva after a fluoride rinse and subsequent sucrose administration. Caries Res. 2002, 36, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, M.; Bartholmes, P. Purification, characterization and inhibition by fluoride of enolase from Streptococcus mutans DSM 320523. Caries Res. 1992, 26, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Guha-Chowdhury, N.; Clark, A.G.; Sissons, C.H. Inhibition of purified enolases from oral bacteria by fluoride. Oral Microbiol. Immunol. 1997, 12, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Assev, S.; Rölla, G. Evidence for presence of a xylitol phosphotransferase system in Streptococcus mutans OMZ 176. Acta Pathol. Microbiol. Immunol. Scand. B 1984, 92, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Trahan, L. Xylitol: A review of its action on mutans streptococci and dental plaque—Its clinical significance. Int. Dent. J. 1995, 45 (Suppl. 1), 77S–92S. [Google Scholar]
- Pihlanto-Leppälä, A.; Söderling, E.; Mäkinen, K.K. Expulsion mechanism of xylitol 5-phosphate in Streptococcus mutans. Scand. J. Dent. Res. 1990, 98, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Trahan, L.; Néron, S.; Bareil, M. Intracellular xylitol-phosphate hydrolysis and efflux of xylitol in Streptococcus sobrinus. Oral Microbiol. Immunol. 1991, 6, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Murakami, Y.; Kubo, S.; Tamori, A.; Itami, S.; Kawamura, E.; Iwaisako, K.; Ikeda, K.; Kawada, N.; Ochiya, T.; Taguchi, Y.H. Comprehensive analysis of transcriptome and metabolome analysis in intrahepatic cholangiocarcinoma and hepatocellular carcinoma. Sci. Rep. 2015, 5, 16294. [Google Scholar] [CrossRef] [PubMed]
- Kami, K.; Fujimori, T.; Sato, H.; Sato, M.; Yamamoto, H.; Ohashi, Y.; Sugiyama, N.; Ishihama, Y.; Onozuka, H.; Ochiai, A.; et al. Metabolomic profiling of lung and prostate tumor tissues by capillary electrophoresis time-of-flight mass spectrometry. Metabolomics 2013, 9, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Urakami, K.; Zangiacomi, V.; Yamaguchi, K.; Kusuhara, M. Impact of 2-deoxy-d-glucose on the target metabolome profile of a human endometrial cancer cell line. Biomed. Res. 2013, 34, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, K.; Fukuda, T.; Isogai, H.; Okumura, K.; Krstic-Demonacos, M.; Isogai, E. Antimicrobial peptide FF/CAP18 induces apoptotic cell death in HCT116 colon cancer cells via changes in the metabolic profile. Int. J. Oncol. 2015, 46, 1516–1526. [Google Scholar] [CrossRef] [PubMed]
- Ohka, F.; Ito, M.; Ranjit, M.; Senga, T.; Motomura, A.; Motomura, K.; Saito, K.; Kato, K.; Kato, Y.; Wakabayashi, T.; et al. Quantitative metabolome analysis profiles activation of glutaminolysis in glioma with IDH1 mutation. Tumour Biol. 2014, 35, 5911–5920. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Washio, J.; Takahashi, T.; Echigo, S.; Takahashi, N. Glucose and glutamine metabolism in oral squamous cell carcinoma: Insight from a quantitative metabolomic approach. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2014, 118, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Miwa, I.; Okudo, J.; Maeda, K.; Okuda, G. Mutarotase effect on colorimetric determination of blood glucose with -D-glucose oxidase. Clin. Chim. Acta 1972, 37, 538–540. [Google Scholar] [CrossRef]
- Sugimoto, M.; Wong, D.T.; Hirayama, A.; Soga, T.; Tomita, M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics 2010, 6, 78–95. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Gao, P.; Wang, X.; Duan, Y. The early diagnosis and monitoring of squamous cell carcinoma via saliva metabolomics. Sci. Rep. 2014, 30. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Wang, D.; Soga, T.; Tomita, M. Salivary Metabolomics for Oral Cancer Detection. In Oral Cancer: Causes, Diagnosis and Treatment; Harris, M.K., Ed.; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2011; pp. 271–278. [Google Scholar]
- Kageyama, G.; Saegusa, J.; Irino, Y.; Tanaka, S.; Tsuda, K.; Takahashi, S.; Sendo, S.; Morinobu, A. Metabolomics analysis of saliva from patients with primary Sjögren’s syndrome. Clin. Exp. Immunol. 2015, 182, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Saruta, J.; Matsuki, C.; To, M.; Onuma, H.; Kaneko, M.; Soga, T.; Tomita, M.; Tsukinoki, K. Physiological and environmental parameters associated with mass spectrometry-based salivary metabolomics profiles. Metabolomics 2013, 9, 454–463. [Google Scholar] [CrossRef]



| Metabolites | Concentration (nmol/mg Wet Weight of Plaque) | ||
|---|---|---|---|
| Before | After | ||
| EMP pathway | |||
| G6P | 0.133 ± 0.032 # | 0.442 ± 0.087 * | |
| F6P | 0.033 ± 0.006 | 0.108 ± 0.025 * | |
| F1,6BP | 0.024 ± 0.010 | 0.099 ± 0.083 | |
| DHAP | 0.037 ± 0.004 | 0.074 ± 0.011 * | |
| 3PG | 0.245 ± 0.165 | 0.218 ± 0.159 | |
| PEP | 0.094 ± 0.035 | 0.062 ± 0.036 | |
| Pyr | 0.588 ± 0.461 | 4.235 ± 2.731 | |
| Lactate | 1.737 ± 0.823 | 13.12 ± 12.71 | |
| PP pathway | |||
| 6PG | 0.008 ± 0.002 | 0.033 ± 0.017 | |
| Ribu 5P | 0.029 ± 0.014 | 0.054 ± 0.021 * | |
| Ribo 5P | 0.011 ± 0.007 | 0.036 ± 0.027 | |
| S7P | 0.058 ± 0.019 | 0.143 ± 0.041 * | |
| E4P | ND | ND | |
| TCA cycle | |||
| ACoA | 0.020 ± 0.006 | 0.045 ± 0.019 | |
| CA | 0.038 ± 0.031 | 0.017 ± 0.006 | |
| AC | 0.001 ± 0.003 | 0.000 ± 0.001 | |
| iCA | 0.001 ± 0.002 | 0.000 ± 0.000 | |
| 2OG | 0.013 ± 0.013 | 0.023 ± 0.012 | |
| SCoA | 0.011 ± 0.018 | 0.019 ± 0.022 | |
| Suc | 1.834 ± 1.320 | 1.650 ± 1.001 | |
| Fum | 0.034 ± 0.039 | 0.018 ± 0.019 | |
| Mal | 0.105 ± 0.059 | 0.074 ± 0.044 | |
| Metabolites | The Rate of Changes in Levels of Metabolites (Times) | |||
|---|---|---|---|---|
| Xylitol | 225 ppm F | 900 ppm F | ||
| EMP pathway | ||||
| G6P | 1.04 ± 0.56 # | 1.40 ± 0.32 * | 1.76 ± 0.60 | |
| F6P | 0.81 ± 0.79 | 3.39 ± 4.78 | 2.18 ± 1.03 | |
| F1,6BP | 0.79 ± 0.80 | 1.64 ± 0.64 | 4.27 ± 2.70 | |
| DHAP | 0.87 ± 0.40 | 0.57 ± 0.45 | 0.44 ± 0.28 | |
| 3PG | 0.94 ± 0.70 | 3.57 ± 2.46 | 9.22 ± 5.22 | |
| PEP | 0.95 ± 0.80 | 0.83 ± 0.83 | 0.57 ± 0.63 | |
| Pyr | 0.82 ± 0.59 | 0.41 ± 0.19 | 0.29 ± 0.15 | |
| Lactate | 1.02 ± 0.55 | 0.70 ± 0.21 | 0.59 ± 0.31 | |
| PP pathway | ||||
| 6PG | 0.97 ± 0.67 | 2.09 ± 0.99 | 4.40 ± 2.48 | |
| Ribu 5P | 0.91 ± 0.40 | 0.80 ± 0.33 | 0.81 ± 0.21 | |
| Ribo 5P | 0.83 ± 0.37 | 0.39 ± 0.45 | 0.28 ± 0.14 | |
| S7P | 0.92 ± 0.45 | 1.18 ± 0.23 | 2.90 ± 3.10 | |
| E4P | ND | ND | ND | |
| Oral Rinse with; | Concentration (nmol/mg Wet Weight of Plaque) |
|---|---|
| Glucose | ND |
| Xylitol + Glucose | 13.9 ± 5.2 # |
| Metabolites | Concentration (nmol/mg Wet Weight of Tissue) | ||
|---|---|---|---|
| Cancer | Normal | ||
| Glucose | 0.84 ± 0.67 #,* | 1.59 ± 1.31 | |
| EMP Pathway | |||
| 3PG | 0.38 ± 0.49 * | 1.64 ± 3.81 | |
| 2PG | 0.06 ± 0.07 * | 0.24 ± 0.54 | |
| Lactate | 76.7 ± 67.1 * | 51.9 ± 43.8 | |
| TCA cycle | |||
| Fum | 0.67 ± 0.36 * | 0.52 ± 0.33 | |
| Mal | 1.83 ± 1.36 * | 1.32 ± 0.88 | |
| The amino acids and the metabolites related to amino acids metabolism | |||
| Glutamine | 9.35 ± 7.53 $ | 11.0 ± 6.63 | |
| Glutamate | 13.8 ± 10.1 * | 9.92 ± 9.52 | |
| Glycine | 8.37 ± 7.94 * | 6.05 ± 6.17 | |
| Aspartate | 5.00 ± 4.69 * | 2.93 ± 3.54 | |
| Proline | 2.92 ± 3.29 * | 2.11 ± 2.35 | |
| Cysteine | 0.21 ± 0.21 * | 0.12 ± 0.14 | |
| Hydroxyproline | 0.34 ± 0.79 * | 0.23 ± 0.49 | |
| Creatine | 8.48 ± 9.39 * | 14.0 ± 12.9 | |
| Creatinine | 0.30 ± 0.47 * | 0.30 ± 0.27 | |
| Putrescine | 0.27 ± 1.07 * | 0.08 ± 0.29 | |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Washio, J.; Takahashi, N. Metabolomic Studies of Oral Biofilm, Oral Cancer, and Beyond. Int. J. Mol. Sci. 2016, 17, 870. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17060870
Washio J, Takahashi N. Metabolomic Studies of Oral Biofilm, Oral Cancer, and Beyond. International Journal of Molecular Sciences. 2016; 17(6):870. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17060870
Chicago/Turabian StyleWashio, Jumpei, and Nobuhiro Takahashi. 2016. "Metabolomic Studies of Oral Biofilm, Oral Cancer, and Beyond" International Journal of Molecular Sciences 17, no. 6: 870. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17060870