Effect of Antimicrobial Denture Base Resin on Multi-Species Biofilm Formation
Abstract
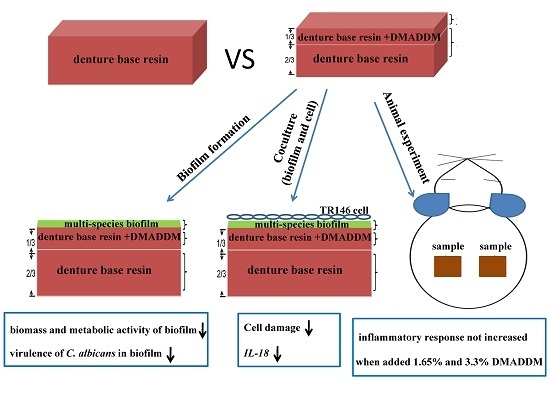
:1. Introduction
2. Results
2.1. Dimethylaminododecyl Methacrylate (DMADDM) Reduced the Viable Microbes in the Multi-Species Biofilms
2.2. DMADDM Changed the Multi-Biofilm Structuel and Inhibited Themetabolic Abilities
2.3. DMADDM Inhibited the Hypal Development and Virulence Ralated Genes Expression of C. albicans in Multi-Species Biofilms
2.4. DMADDM Reduced the Cell Damage Caused by Multi-Species Biofilm and Also Redeced the Candida Receptor and the Pro-Flammatory Cytokine Prodction of TR-146 Cell
2.5. Biocompatibility of DMADDM Modified Denture Base Resin in Vivo
3. Discussion
4. Materials and Methods
4.1. DMADDM Synthesis and Preparation of Specimen
4.2. Microbial Cultures and Multi-Species Biofilm Formation
4.3. pH Measurement
4.4. Colony-Forming Unit (CFU) Counts
4.5. 2,3-Bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT) Array
4.6. Cell Culture
4.7. Cell Damage Assay
4.8. RNA Isolation and Real-Time PCR
4.9. Biofilm Imaging
4.10. Biocompatibility Experiment
4.11. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| DMADDM | Dimethylaminododecyl methacrylate |
| QAS | Quaternary ammonium salts |
| DMAD | 1-(dimethylamino)dodecan |
| BEMA | 2-bromoethyl methacrylate |
| CLSM | Confocal laser scanning microscope |
| LDH | Lactate dehydrogenase |
Appendix A
Synthesis of DMADDM
Appendix B

Appendix C

Appendix D
| Primers | Nucelotide Sequence (5′–3′) |
|---|---|
| ACT1 | FW-TGCTGAACGTATGCAAAAGG |
| RV-TGAACAATGGATGGACCAGA | |
| ALS1 | FW-CCCAACTTGGAATGCTGTTT |
| RV-TTTCAAAGCGTCGTTCACAG | |
| ALS3 | FW-CTGGACCACCAGGAAACACT |
| RV-GGTGGAGCGGTGACAGTAGT | |
| SAP4 | FW-GTCAATGTCAACGCTGGTGTCC |
| RV-ATTCCGAAGCAGGAACGGTGTCC | |
| SAP6 | FW-AAAATGGCGTGGTGACAGAGGT |
| RV-CGTTGGCTTGGAAACCAATACC | |
| HWP1 | FW-TCTACTGCTCCAGCCACTGA |
| RV-CCAGCAGGAATTGTTTCCAT | |
| PLD1 | FW-GCCAAGAGAGCAAGGGTTAGCA |
| RV-CGGATTCGTCATCCATTTCTCC | |
| β-actin | FW-GAGCACAGAGCCTCGCCTTTGCCGAT |
| RV-ATCCTTCTGACCCATGCCCACCATCACG | |
| IL-18 | FW-CCTTCCAGATCGCTTCCTCTCGCAACAA |
| RV-CAAGCTTGCCAAAGTAATCTGATTCCAGGT | |
| Dectin1 | FW-ACAGCAATGAGGCGCCAAGGAGGAGATG |
| RV-GGAGCAGAAAGAAAAGAGCTCCCAAATGCT |
References
- Nakahara, T.; Harada, A.; Yamada, Y.; Odashima, Y.; Nakamura, K.; Inagaki, R.; Kanno, T.; Sasaki, K.; Niwano, Y. Influence of a new denture cleaning technique based on photolysis of H2O2 the mechanical properties and color change of acrylic denture base resin. Dent. Mater. J. 2013, 32, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Akpan, A.; Morgan, R. Oral candidiasis. Postgrad. Med. J. 2002, 78, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Pereira-Cenci, T.; del bel Cury, A.A.; Crielaard, W.; Ten Cate, J.M. Development of Candida-associated denture stomatitis: New insights. J. Appl. Oral Sci. 2008, 16, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Arendorf, T.M.; Walker, D.M. Denture stomatitis: A review. J. Oral Rehabil. 1987, 14, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Nikawa, H.; Hamada, T.; Yamamoto, T. Denture plaque—Past and recent concerns. J. Dent. 1998, 26, 299–304. [Google Scholar] [CrossRef]
- Jepson, N.J.; Moynihan, P.J.; Kelly, P.J.; Watson, G.W.; Thomason, J.M. Restorative dentistry: Caries incidence following restoration of shortened lower dental arches in a randomized controlled trial. Br. Dent. J. 2001, 191, 140–144. [Google Scholar] [CrossRef] [PubMed]
- Coulthwaite, L.; Verran, J. Potential pathogenic aspects of denture plaque. Br. J. Biomed. Sci. 2007, 64, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Taylor, G.W.; Loesche, W.J.; Terpenning, M.S. Impact of oral diseases on systemic health in the elderly: Diabetes mellitus and aspiration pneumonia. J. Public Health Dent. 2000, 60, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Parahitiyawa, N.B.; Jin, L.J.; Leung, W.K.; Yam, W.C.; Samaranayake, L.P. Microbiology of odontogenic bacteremia: Beyond endocarditis. Clin. Microbiol. Rev. 2009, 22, 46–64. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Finnegan, M.B.; Özkan, S.; Kim, Y.; Lillehoj, P.B.; Ho, C.M.; Lux, R.; Mito, R.; Loewy, Z.; Shi, W. In vitro study of biofilm formation and effectiveness of antimicrobial treatment on various dental material surfaces. Mol. Oral Microbiol. 2010, 25, 384–390. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.; Jagger, R.; Vowles, R.; Moran, J. Comparative stain removal properties of four commercially available denture cleaning products: An in vitro study. Int. J. Dent. Hyg. 2011, 9, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Stopiglia, C.D.; Collares, F.M.; Ogliari, F.A.; Piva, E.; Fortes, C.B.; Samuel, S.M.; Scroferneker, M.L. Antimicrobial activity of [2-(methacryloyloxy)ethyl]trimethylammonium chloride against Candida spp. Rev. Iberoam. Micol. 2012, 29, 20–23. [Google Scholar] [CrossRef] [PubMed]
- Sivakumar, I.; Arunachalam, K.S.; Sajjan, S.; Ramaraju, A.V.; Rao, B.; Kamaraj, B. Incorporation of Antimicrobial Macromolecules in Acrylic Denture Base Resins: A Research Composition and Update. J. Prosthodont. 2014, 23, 284–290. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Torres, L.S.; Mendieta, I.; Nuñez-Anita, R.E.; Cajero-Juárez, M.; Castaño, V.M. Cytocompatible antifungal acrylic resin containing silver nanoparticles for dentures. Int. J. Nanomed. 2012, 7, 4777–4786. [Google Scholar]
- Paleari, A.G.; Marra, J.; Pero, A.C.; Rodriguez, L.S.; Ruvolo-Filho, A.; Compagnoni, M.A. Effect of incorporation of 2-tert-butylaminoethyl methacrylate on flexural strength of a denture base acrylic resin. J. Appl. Oral Sci. 2011, 19, 195–199. [Google Scholar] [PubMed]
- Cao, Z.; Sun, X.; Yeh, C.K.; Sun, Y. Rechargeable infection-responsive antifungal denture materials. J. Dent. Res. 2010, 89, 1517–1521. [Google Scholar] [CrossRef] [PubMed]
- Arai, T.; Ueda, T.; Sugiyama, T.; Sakurai, K. Inhibiting microbial adhesion to denture base acrylic resin by titanium dioxide coating. J. Oral Rehabil. 2009, 36, 902–908. [Google Scholar] [CrossRef] [PubMed]
- Redding, S.; Bhatt, B.; Rawls, H.R.; Siegel, G.; Scott, K.; Lopez-Ribot, J. Inhibition of Candida albicans biofilm formation on denture material. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2009, 107, 669–672. [Google Scholar] [CrossRef] [PubMed]
- Calderone, R.A.; Fonzi, W.A. Virulence factors of Candida albicans. Trends Microbiol. 2001, 9, 327–335. [Google Scholar] [CrossRef]
- Xu, H.; Sobue, T.; Thompson, A.; Xie, Z.; Poon, K.; Ricker, A.; Cervantes, J.; Diaz, P.I.; Dongari-Bagtzoglou, A. Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell. Microbiol. 2014, 16, 214–231. [Google Scholar] [CrossRef] [PubMed]
- Ricker, A.; Vickerman, M.; Dongari-Bagtzoglou, A. Streptococcus gordonii glucosyltransferase promotes biofilm interactions with Candida albicans. J. Oral Microbiol. 2014, 6, 23419. [Google Scholar] [CrossRef] [PubMed]
- Sztajer, H.; Szafranski, S.P.; Tomasch, J.; Reck, M.; Nimtz, M.; Rohde, M.; Wagner-Döbler, I. Cross-feeding and interkingdom communication in dual-species biofilms of Streptococcus mutans and Candida albicans. ISME J. 2014, 8, 2256–2271. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Weir, M.D.; Fouad, A.F.; Xu, H.H. Time-kill behaviour against eight bacterial species and cytotoxicity of antibacterial monomers. J. Dent. 2013, 41, 881–891. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, K.; Zhou, X.; Xu, N.; Xu, H.H.; Weir, M.D.; Ge, Y.; Wang, S.; Li, M.; Li, Y.; Xu, X.; Cheng, L. Antibacterial Effect of Dental Adhesive Containing Dimethylaminododecyl Methacrylate on the Development of Streptococcus mutans Biofilm. Int. J. Mol. Sci. 2014, 15, 12791–12806. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Wang, S.; Zhou, X.; Xu, H.H.; Weir, M.D.; Ge, Y.; Li, M.; Wang, S.; Li, Y.; Xu, X.; Zheng, L.; Cheng, L. Effect of Antibacterial Dental Adhesive on Multispecies Biofilms Formation. J. Dent. Res. 2015, 94, 622–629. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Weir, M.D.; Zhang, K.; Arola, D.D.; Zhou, X.; Xu, H.H. Dental primer and adhesive containing a new antibacterial quaternary ammonium monomer dimethylaminododecyl methacrylate. J. Dent. 2013, 41, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Cheng, L.; Wu, E.J.; Weir, M.D.; Bai, Y.; Xu, H.H. Effect of water-ageing on dentine bond strength and anti-biofilm activity of bonding agent containing new monomer dimethylaminododecyl methacrylate. J. Dent. 2013, 41, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Cai, K.; Fang, J.; Lai, M.; Li, J.; Hou, Y.; Luo, Z.; Hu, Y.; Tang, L. Dual action antibacterial TiO2 nanotubes incorporated with silver nanoparticles and coated with a quaternary ammonium salt (QAS). Surf. Coat. Technol. 2013, 216, 158–165. [Google Scholar] [CrossRef]
- Imazato, S.; Ma, S.; Chen, J.H.; Xu, H.H. Therapeutic polymers for dental adhesives: Loading resins with bio-active components. Dent. Mater. 2014, 30, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Weir, M.D.; Antonucci, J.M.; Schumacher, G.E.; Zhou, X.D.; Xu, H.H. Evaluation of three-dimensional biofilms on antibacterial bonding agents containing novel quaternary ammonium methacrylates. Int. J. Oral Sci. 2014, 6, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Han, Q.; Zhou, X.; Zhang, K.; Wang, S.; Xu, H.H.; Weir, M.D.; Feng, M.; Li, M.; Ren, B.; Cheng, L. Heat-polymerized resin containing dimethylaminododecyl methacrylate inhibits Candida albicans biofilm. J. Dent. 2016. submitted. [Google Scholar]
- Cavalcanti, Y.W.; Morse, D.J.; da Silva, W.J.; Del-Bel-Cury, A.A.; Wei, X.; Wilson, M.; Milward, P.; Lewis, M.; Bradshaw, D.; Williams, D.W. Virulence and pathogenicity of Candida albicans is enhanced in biofilms containing oral bacteria. Biofouling 2015, 31, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Chandra, J.; Mukherjee, P.K.; Leidich, S.D.; Faddoul, F.F.; Hoyer, L.; Douglas, L.J.; Ghannoum, M.A. Antifungal resistance of Candidal biofilms formed on denture acrylic in vitro. J. Dent. Res. 2001, 80, 903–908. [Google Scholar] [CrossRef] [PubMed]
- Braga, P.C.; Culici, M.; Alfieri, M.; dal Sasso, M. Thymol inhibits Candida albicans biofilm formation and mature biofilm. Int. J. Antimicrob. Agents 2008, 31, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Pusateri, C.R.; Monaco, E.A.; Edgerton, M. Sensitivity of Candida albicans biofilm cells grown on denture acrylic to antifungal proteins and chlorhexidine. Arch. Oral Biol. 2009, 54, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Campos, M.S.; Marchini, L.; Bernardes, L.A.; Paulino, L.C.; Nobrega, F.G. Biofilm microbial communities of denture stomatitis. Oral Microbiol. Immunol. 2008, 23, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Shirtliff, M.E.; Peters, B.M.; Jabra-Rizk, M.A. Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol. Lett. 2009, 299, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Salerno, C.; Pascale, M.; Contaldo, M.; Esposito, V.; Busciolano, M.; Milillo, L.; Guida, A.; Petruzzi, M.; Serpico, R. Candida-associated denture stomatitis. Med. Oral Patol. Oral Cir. Bucal 2011, 16, e139–e143. [Google Scholar] [CrossRef] [PubMed]
- Sudbery, P.E. Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 2011, 9, 737–748. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Weir, M.D.; Chen, J.; Xu, H.H. Effect of charge density of bonding agent containing a new quaternary ammonium methacrylate on antibacterial and bonding properties. Dent. Mater. 2014, 30, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wang, P.; Weir, M.D.; Fouad, A.F.; Xu, H.H. Evaluation of antibacterial and remineralizing nanocomposite and adhesive in rat tooth cavity model. Acta Biomater. 2014, 10, 2804–2813. [Google Scholar] [CrossRef] [PubMed]
- McBain, A.J.; Sissons, C.; Ledder, R.G.; Sreenivasan, P.K.; de Vizio, W.; Gilbert, P. Development and characterization of a simple perfused oral microcosm. J. Appl. Microbiol. 2005, 98, 624–634. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.M.; Ward, R.M.; Rane, H.S.; Lee, S.A.; Noverr, M.C. Efficacy of ethanol against Candida albicans and Staphylococcus aureus polymicrobial biofilms. Antimicrob. Agents Chemother. 2013, 57, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Dalle, F.; Wächtler, B.; L’Ollivier, C.; Holland, G.; Bannert, N.; Wilson, D.; Labruère, C.; Bonnin, A.; Hube, B. Cellular interactions of Candida albicans with human oral epithelial cells and enterocytes. Cell. Microbiol. 2010, 12, 248–271. [Google Scholar] [CrossRef] [PubMed]
- Boros-Majewska, J.; Salewska, N.; Borowski, E.; Milewski, S.; Malic, S.; Wei, X.Q.; Hayes, A.J.; Wilson, M.J.; Williams, D.W. Novel Nystatin A1 derivatives exhibiting low host cell toxicity and antifungal activity in an in vitro model of oral candidosis. Med. Microbiol. Immunol. 2014, 203, 341–355. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Klein, M.I.; Falsetta, M.L.; Lu, B.; Delahunty, C.M.; Yates, J.R., 3rd; Heydorn, A.; Koo, H. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog. 2012, 8, e1002623. [Google Scholar] [CrossRef] [PubMed]
- Yudovin-Farber, I.; Beyth, N.; Nyska, A.; Weiss, E.I.; Golenser, J.; Domb, A.J. Surface characterization and biocompatibility of restorative resin containing nanoparticles. Biomacromolecules 2008, 9, 3044–3050. [Google Scholar] [CrossRef] [PubMed]





© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, K.; Ren, B.; Zhou, X.; Xu, H.H.K.; Chen, Y.; Han, Q.; Li, B.; Weir, M.D.; Li, M.; Feng, M.; et al. Effect of Antimicrobial Denture Base Resin on Multi-Species Biofilm Formation. Int. J. Mol. Sci. 2016, 17, 1033. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17071033
Zhang K, Ren B, Zhou X, Xu HHK, Chen Y, Han Q, Li B, Weir MD, Li M, Feng M, et al. Effect of Antimicrobial Denture Base Resin on Multi-Species Biofilm Formation. International Journal of Molecular Sciences. 2016; 17(7):1033. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17071033
Chicago/Turabian StyleZhang, Keke, Biao Ren, Xuedong Zhou, Hockin H. K. Xu, Yu Chen, Qi Han, Bolei Li, Michael D. Weir, Mingyun Li, Mingye Feng, and et al. 2016. "Effect of Antimicrobial Denture Base Resin on Multi-Species Biofilm Formation" International Journal of Molecular Sciences 17, no. 7: 1033. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17071033