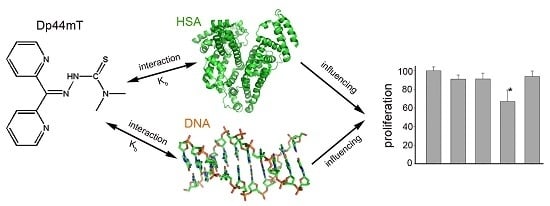
Analysis of the Interaction of Dp44mT with Human Serum Albumin and Calf Thymus DNA Using Molecular Docking and Spectroscopic Techniques
Abstract
:1. Introduction
2. Results and Discussion
2.1. Human Serum Albumin (HSA) Affected the Cytotoxicity of Dp44mT and Its Copper Complex (Dp44mT-Cu)
2.2. UV-VIS Spectral Study of the Interaction between Dp44mT and HSA
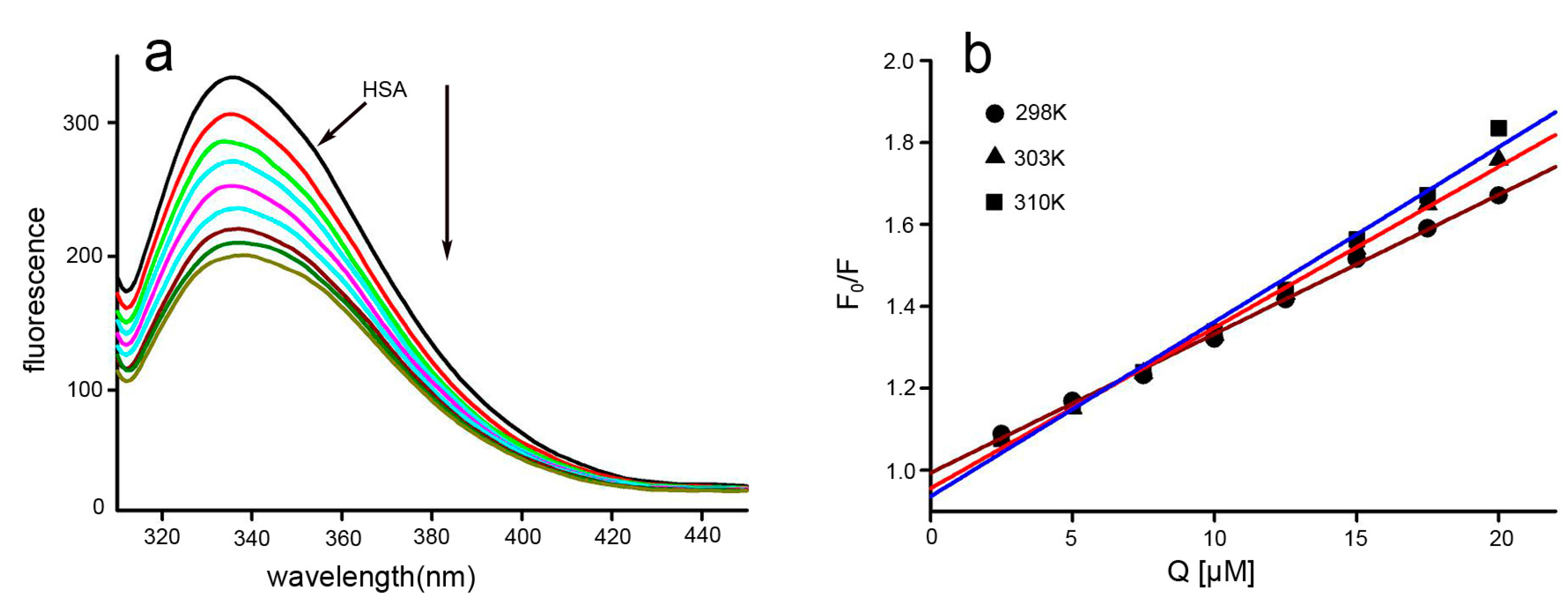
2.3. Fluorescence Quenching and Quenching Mechanism Analysis
2.4. Binding Constants and Binding Sites
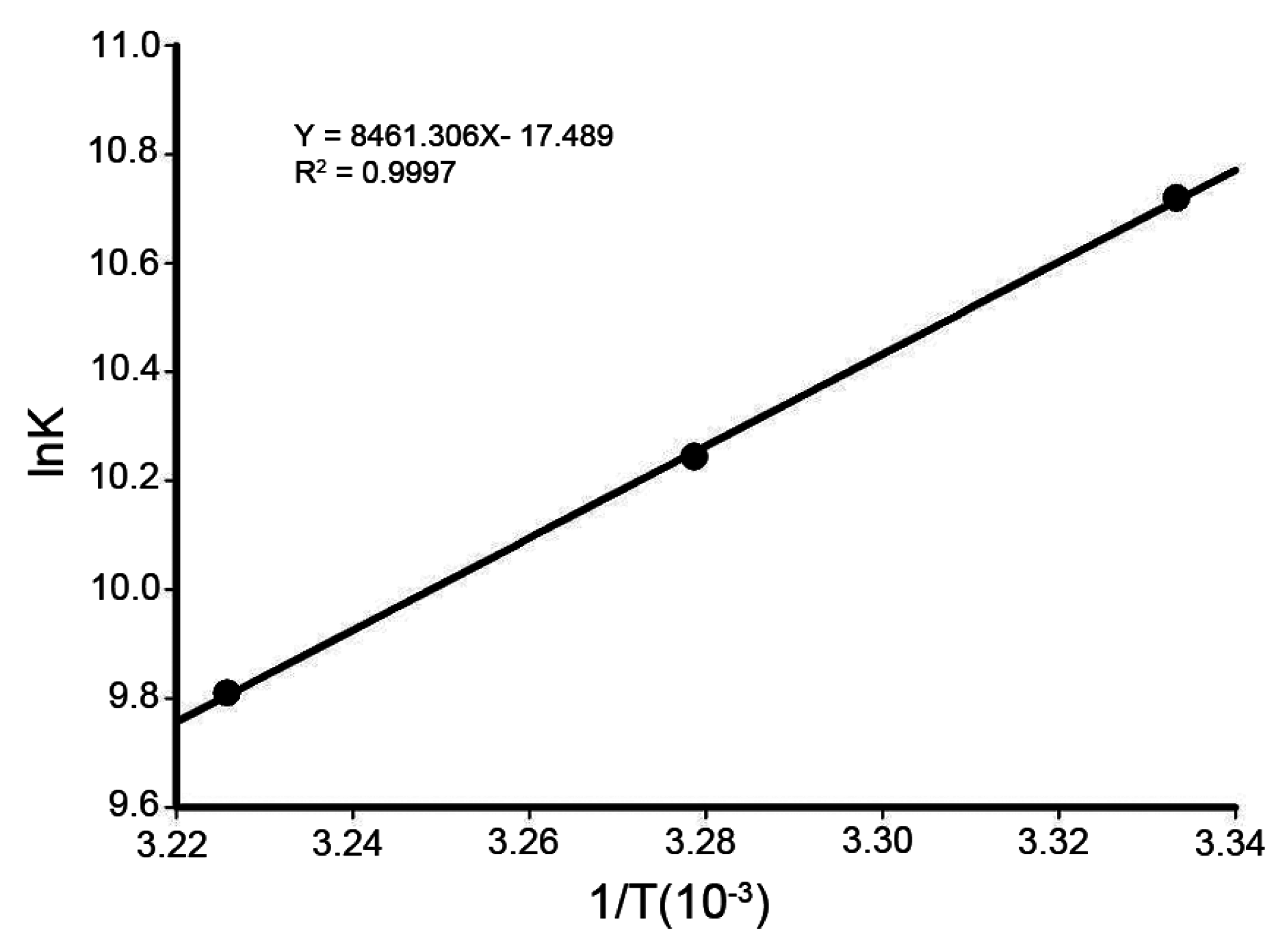
2.5. Thermodynamic Parameters and Nature of the Binding Forces
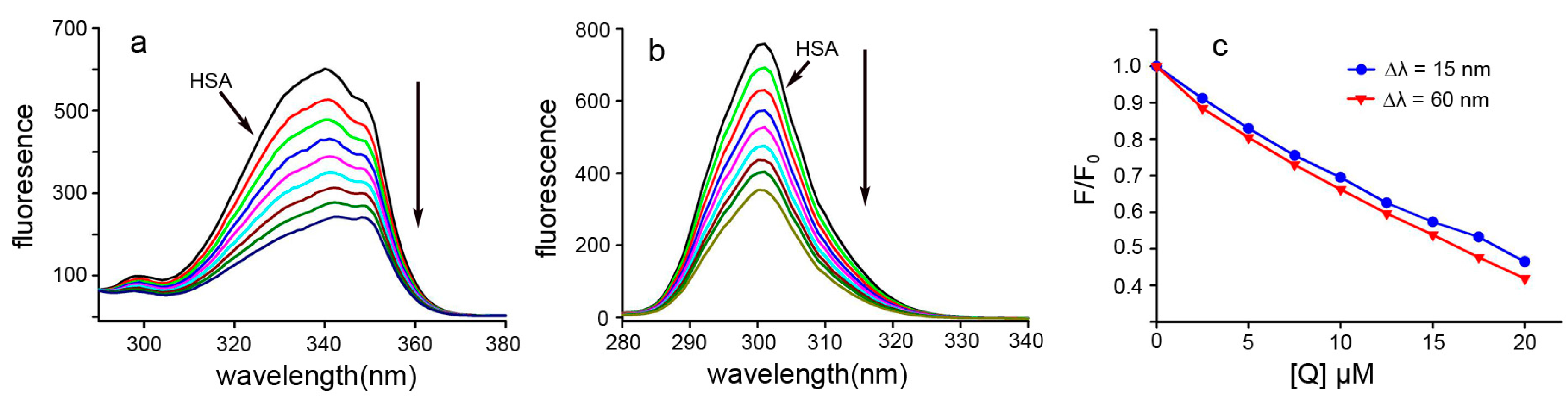
2.6. Energy Transfer between Dp44mT and HSA
2.7. Conformational Investigation by Synchronous Fluorescence
2.8. Circular Dichroism (CD) Spectra
2.9. Interaction of Dp44mT with Ct-DNA
2.10. Molecular Docking
3. Experimental Section
3.1. Materials
3.2. Proliferation Assay
3.3. Spectrophotometric Study
3.4. Fluorescence Study
3.5. Measurement of CD Spectra
3.6. Molecular Docking
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| HSA | Human Serum Albumin |
| Dp44mT | Di-2-pyridylketone-4,4,-dimethyl-3-thiosemicarbazone |
| FRET | Fluorescence Resonance Energy Transfer |
| CD | Circular Dichroism |
| Tf | Transferrin |
References
- Pahl, P.M.; Horwitz, L.D. Cell permeable iron chelators as potential cancer chemotherapeutic agents. Cancer Investig. 2005, 23, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Becker, E.M.; Lovejoy, D.B.; Greer, J.M.; Watts, R.; Richardson, D.R. Identification of the di-pyridyl ketone isonicotinoyl hydrazone (PKIH) analogues as potent iron chelators and anti-tumour agents. Br. J. Pharmacol. 2003, 38, 819–830. [Google Scholar] [CrossRef] [PubMed]
- Chaston, T.B.; Lovejoy, D.B.; Watts, R.N.; Richardson, D.R. Examination of the anti-proliferative activity of iron chelators. Clin. Cancer Res. 2003, 9, 402–414. [Google Scholar] [PubMed]
- Whitnall, M.; Howard, J.; Ponka, P.; Richardson, D.R. A class of iron chelators with a wide spectrum of potent antitumor activity that overcomes resistance to chemotherapeutics. Proc. Natl. Acad. Sci. USA 2006, 103, 14901–14906. [Google Scholar] [CrossRef] [PubMed]
- Kovacevic, Z.; Chikhani, S.; Lovejoy, D.B.; Richardson, D.R. Novel thiosemicarbazone iron chelators induce up-regulation and phosphorylation of the metastasis suppressor N-myc down-stream regulated gene 1: A new strategy for the treatment of pancreatic cancer. Mol. Pharmacol. 2011, 80, 598–609. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Xing, F.; Iiizumi-Gairani, M.; Okuda, H.; Watabe, M.; Pai, S.K.; Pandey, P.R.; Hirota, S.; Kobayashi, A.; Mo, Y.Y.; et al. N-myc downstream regulated gene 1 modulates Wnt-β-catenin signalling and pleiotropically suppresses metastasis. EMBO Mol. Med. 2012, 4, 93–108. [Google Scholar] [CrossRef] [PubMed]
- Rao, V.A.; Klein, S.R.; Agama, K.K.; Toyoda, E.; Adachi, N.; Pommier, Y.; Shacter, E.B. The iron chelator Dp44mT causes DNA damage and selective inhibition of topoisomerase IIA in breast cancer cells. Cancer Res. 2009, 69, 948–957. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Lovejoy, D.B.; Richardson, D.R. Novel di-2-pyridyl-derived iron chelators with marked and selective antitumor activity: In vitro and in vivo assessment. Blood 2004, 104, 1450–1458. [Google Scholar] [CrossRef] [PubMed]
- Kovacevic, Z.; Sivagurunathan, S.; Mangs, H.; Chikhani, S.; Zhang, D.; Richardson, D.R. The metastasis suppressor, N-myc downstream regulated gene 1 (NDRG1), upregulates p21 via p53-independent mechanisms. Carcinogenesis 2011, 32, 732–740. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.J.R.; Mills, T.M.; Shafie, N.H.; Merlot, A.M.; Moussa, R.S.; Kalinowski, D.S.; Kovacevic, Z.; Richardson, D.R. Expanding horizons in iron chelation and the treatment of cancer: Role of iron in the regulation of ER stress and the epithelial-mesenchymal transition. Biochim. Biophys. Acta 2014, 1845, 166–181. [Google Scholar] [CrossRef] [PubMed]
- Flarakos, J.; Morand, K.L.; Vouros, P. High-throughput solution-based medicinal library screening against human serum albumin. Anal. Chem. 2005, 77, 1345–1353. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Zhang, L.; Sun, Y.; Diao, J.X.; Yang, X.L.; Sun, Y.; Zhang, L. Specificity and affinity of phenosafranine protein adduct: Insights from biophysical aspects. J. Lumin. 2012, 132, 629–635. [Google Scholar] [CrossRef]
- Merlot, A.M.; Sahni, S.; Lane, D.J.R.; Fordham, A.M.; Pantarat, N.; Hibbs, D.E.; Richardson, V.; Doddareddy, M.R.; Jennifer, A.; Huang, M.L.H.; et al. Potentiating the cellular targeting and anti-tumor activity of Dp44mT via binding to human serum albumin: Two saturable mechanisms of Dp44mT uptake by cells. Oncotarget 2015, 6, 10374–10398. [Google Scholar] [CrossRef] [PubMed]
- Równicka-Zubik, J.; Sułkowski, L.; Toborek, M. Interactions of PCBs with human serum albumin: In vitro spectroscopic study. Spectrochim. Acta Part A 2014, 124, 632–637. [Google Scholar] [CrossRef] [PubMed]
- Sirajuddin, M.; Ali, S.; Badshah, A. Drug-DNA interactions and their study by UV-Visible, fluorescence spectroscopies and cyclic voltametry. J. Photochem. Photobiol. B 2013, 124, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Li, C.P.; Huang, T.F.; Fu, Y.; Liu, Y.X.; Zhou, S.F.; Qi, Z.Y.; Li, C.Z. Interaction of Di-2-pyridylketone 2-pyridine carboxylic acid hydrazone and its copper complex with BSA: Effect on antitumor activity as revealed by spectroscopic studies. Molecules 2016. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.Y.; Xi, X.L.; Yang, P. Studies on DNA binding to metal complexes of Sal2trien. Biochemistry 2007, 72, 37–43. [Google Scholar] [CrossRef]
- KuntzJr, I.D.; Gasparro, F.P.; JohnstonJr, M.D.; Taylor, R.P. Molecular interactions and the Benesi-Hildebrand equation. J. Am. Chem. Soc. 1968, 90, 4778–4781. [Google Scholar] [CrossRef]
- Gorza, F.D.S.; Pedro, G.C.; Trescher, T.F.; da Silva, R.J.; Silva, J.R.; de Souza, N.C. Morphological analysis and interaction of chlorophyll and BSA. BioMed Res. Int. 2014. Available online: http://www.hindawi.com/journals/bmri/2014/872701/abs/ (accessed on 18 May 2014).
- Naveenraj, S.; Anandan, S. Binding of serum albumins with bioactive substances—Nanoparticles to drugs. J. Photochem. Photobiol. C 2013, 14, 53–71. [Google Scholar] [CrossRef]
- Li, D.J.; Zhu, M.; Xu, C.; Chen, J.J.; Ji, B.M. The effect of Cu2+ or Fe3+ on the noncovalent binding of rutin with bovine serum albumin by spectroscopic analysis. Spectrochim. Acta A 2011, 78, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.L.; Yao, N.N.; Xu, H.R.; Wang, T.S.; Li, G.Y.; Li, Z.Q. Characterization of the Interaction between Eupatorin and Bovine Serum Albumin by Spectroscopic and Molecular Modeling Methods. Int. J. Mol. Sci. 2013, 14, 14185–14203. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R.; Weber, G. Quenching of fluorescence by oxygen. Probe for structural fluctuations in macromolecules. Biochemistry 1973, 12, 4161–4170. [Google Scholar] [CrossRef] [PubMed]
- Bijari, N.; Shokoohini, Y.; Ashrafi-Kooshk, M.R.; Ranjbara, S.; Parvaneh, S.; Moieni-Arya, M.; Khodarahmi, R. Spectroscopic study of interaction between osthole and human serum albumin: Identification of possible binding site of the compound. J. Lumin. 2013, 143, 328–336. [Google Scholar] [CrossRef]
- Ahmed, M.; Guleria, A.; Singh, A.K.; Bandyopadhyry, T.; Sarkar, S. Deciphering the binding modes of hematoporphyrin to bovine serum albumin. Indian J. Biochem. Biophys. 2014, 51, 175–187. [Google Scholar] [PubMed]
- Li, D.J.; Zhu, J.F.; Jin, J.; Yao, X.J. Studies on the binding of nevadensin to human serum albumin by molecular spectroscopy and modeling. J. Mol. Struct. 2007, 846, 34–41. [Google Scholar] [CrossRef]
- Timaseff, S.N. Thermodynamics of Protein Interactions. In Proteins of Biological Fluids; Pergamon Press: Oxford, UK, 1972; pp. 511–519. [Google Scholar]
- Mohammadi, F.; Moeeni, M. Study on the interactions of trans-resveratrol and curcumin with bovine α-lactalbumin by spectroscopic analysis and molecular docking. Mater. Sci. Eng. 2015, 50, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.; Khan, A.Y.; Kumar, G.S. Interaction of the anticancer plant alkaloid sanguinarine with bovine serum albumin. PLoS ONE 2011. [Google Scholar] [CrossRef] [PubMed]
- Varlan, A.; Hillebrand, M. Bovine and human serum albumin interactions with 3-carboxyphenoxathiin studied by fluorescence and circular dichroism spectroscopy. Molecules 2010, 15, 3905–3919. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.X.; Liang, W.J.; Luo, X.J.; Gu, Y.Q. Fluorescence spectroscopy study on the interaction between evodiamine and bovine serum albumin. J. Chem. 2013. [Google Scholar] [CrossRef]
- Ibrahim, N.; Ibrahim, H.; Kim, S.; Nallet, J.P.; Nepveu, F. Interactions between antimalarial indolone-N-oxide derivatives and human serum albumin. Biomacromolecules 2010, 11, 3341–3351. [Google Scholar] [CrossRef] [PubMed]
- Sułkowska, A. Interaction of drugs with bovine and human serum albumin. J. Mol. Struct. 2002, 614, 227–232. [Google Scholar] [CrossRef]
- Suryawanshi, V.D.; Anbhule, P.V.; Gore, A.H.; Patil, S.R.; Kolekar, G.B. A spectral deciphering the perturbation of model transporter protein (HSA) by antibacterial pyrimidine derivative: Pharmacokinetic and biophysical insights. J. Photochem. Photobiol. B 2013, 118, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.; Weljie, A.M.; Vogel, H.J. Tryptophan fluorescence quenching by methionine and selenomethionine residues of calmodulin: Orientation of peptide and protein binding. Biochemistry 1998, 37, 3187–3195. [Google Scholar] [CrossRef] [PubMed]
- Kandagal, P.B.; Ashoka, S.; Seetharamappa, J.; Shaikh, S.M.T.; Jadegoud, Y.; Ijare, O.B. Study of the interaction of an anticancer drug with human and bovine serum albumin: Spectroscopic approach. J. Pharm. Biomed. Anal. 2006, 41, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Perez-Iratxeta, C.; Andrade-Navarro, M.A. K2D2: Estimation of protein secondary structure from circular dichroism spectra. BMC Struct. Biol. 2008. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.E.; Zhao, Y.K.; Xu, Y.Y.; Li, C.Z.; Hu, Z.D.; Xu, Q.H. Studies on the fluorescence reaction between nucleic acid and the complex of cobalt(II) with 5-(3-fluoro-4-chlorophenylazo)-8-sulfonamidoquinoline and its applications. Anal. Biochem. 2000, 277, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Zsila, F.; Subdomain, I.B. Is the third major drug binding region of human serum albumin: Toward the three-sites model. Mol. Pharm. 2013, 10, 1668–1682. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Petitpas, I.; Petersen, C.E.; Ha, C.E.; Bhattacharya, A.A.; Zunszain, P.A.; Ghuman, J.; Bhagavan, N.V.; Curry, S. Structural basis of albumin-thyroxine interactions and familial dysalbuminemic hyperthyroxinemia. Proc. Natl. Acad. Sci. USA 2003, 100, 6440–6445. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System; DeLano Scientific: San Carlos, CA, USA, 2002; Available online: http://www.pymol.org (accessed on 5 July 2012).
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]







| pH | T (K) | Ksv (104 M−1) | Kq (1012 M−1) | R2 |
|---|---|---|---|---|
| 7.4 | 290 | 3.39 | 3.39 | 0.9988 |
| 300 | 3.92 | 3.92 | 0.9962 | |
| 310 | 4.26 | 4.26 | 0.9922 |
| pH | T (K) | Ksv (104 M−1) | R2 |
|---|---|---|---|
| 7.4 | 298 | 4.52 | 0.997 |
| 303 | 2.81 | 0.997 | |
| 310 | 1.82 | 0.999 |
| pH | T (K) | Ka (104 M−1) | n | R2 |
|---|---|---|---|---|
| 7.4 | 298 | 2.82 | 1.112 | 0.997 |
| 303 | 9.68 | 1.027 | 0.992 | |
| 310 | 17.46 | 1.019 | 0.995 |
| pH | T (K) | ΔG (kJ/mol) | ΔH (kJ/mol) | ΔS (J/mol·K) |
|---|---|---|---|---|
| 7.4 | 298 | −27.01 | −70.34 | −145.41 |
| 300 | −26.28 | |||
| 310 | −25.26 |
| System | J (cm3·L·mol−1) | E (%) | R0 (nm) | r (nm) |
|---|---|---|---|---|
| HSA-Dp44mT | 8.38×1013 | 0.341 | 2.40 | 2.06 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Z.; Liu, Y.; Zhou, S.; Fu, Y.; Li, C. Analysis of the Interaction of Dp44mT with Human Serum Albumin and Calf Thymus DNA Using Molecular Docking and Spectroscopic Techniques. Int. J. Mol. Sci. 2016, 17, 1042. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17071042
Xu Z, Liu Y, Zhou S, Fu Y, Li C. Analysis of the Interaction of Dp44mT with Human Serum Albumin and Calf Thymus DNA Using Molecular Docking and Spectroscopic Techniques. International Journal of Molecular Sciences. 2016; 17(7):1042. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17071042
Chicago/Turabian StyleXu, Zhongjie, Youxun Liu, Sufeng Zhou, Yun Fu, and Changzheng Li. 2016. "Analysis of the Interaction of Dp44mT with Human Serum Albumin and Calf Thymus DNA Using Molecular Docking and Spectroscopic Techniques" International Journal of Molecular Sciences 17, no. 7: 1042. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17071042