A Review of Analytical Techniques and Their Application in Disease Diagnosis in Breathomics and Salivaomics Research
Abstract
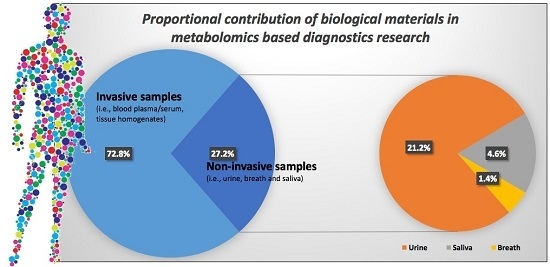
:1. Introduction
2. Breath and Saliva Sampling Considerations
3. Sampling Techniques
3.1. Saliva Sampling
3.2. Breath Sampling
4. Instrumentation and Analytical Techniques
4.1. Nuclear Magnetic Resonance (NMR) Spectroscopy
4.2. Gas and Liquid Chromatography Mass Spectrometry (GC-MS and LC-MS)
4.3. Capillary Electrophoresis
4.4. NMR-GC Hyphenation
4.5. Direct Injection Mass Spectrometry
4.6. Ion-Mobility Spectrometry
4.7. Ultra Violet and Infra Red Spectroscopy
4.8. Electronic Noses
4.9. Data Processing and Statitical Analysis
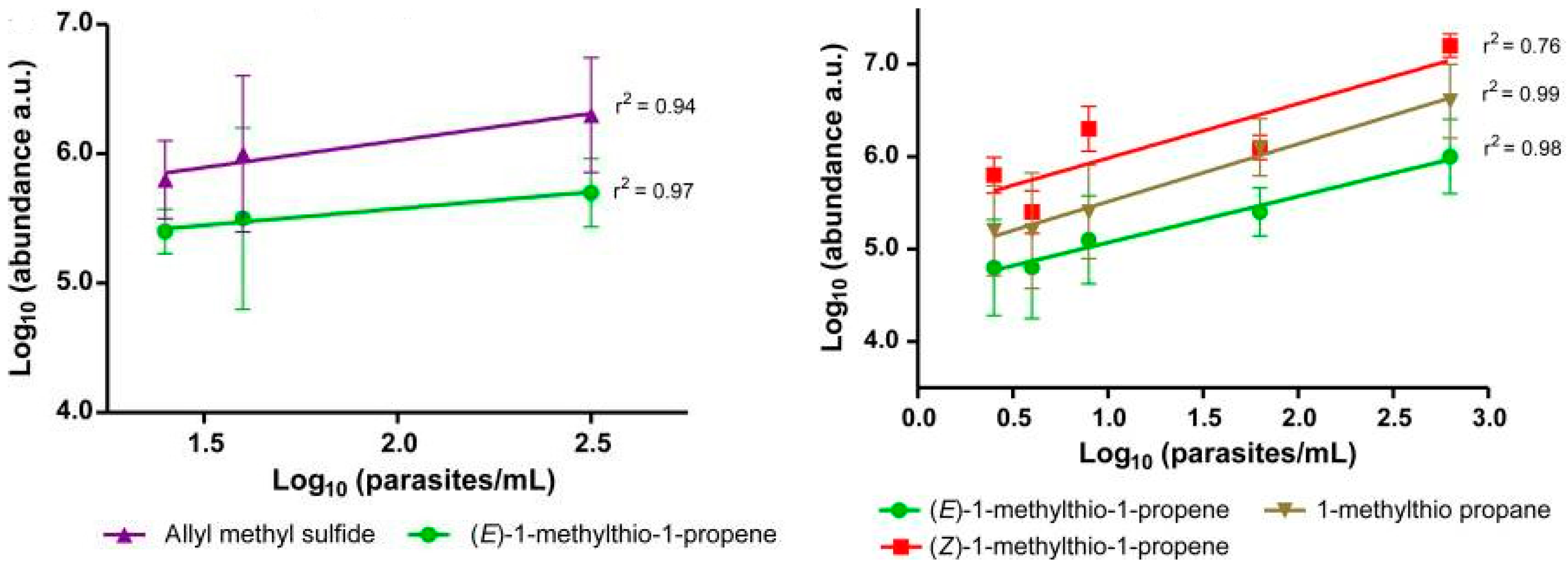
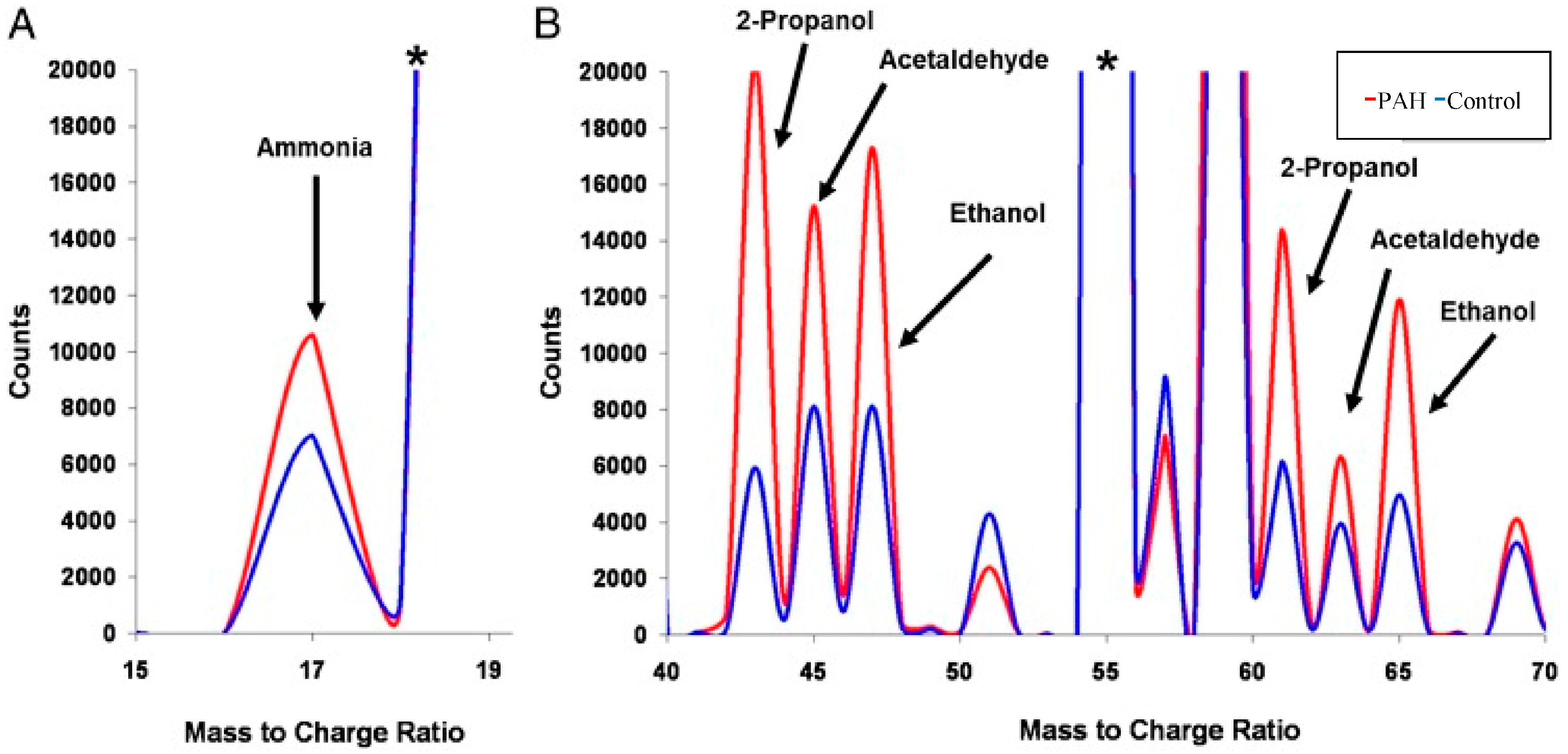
5. Breathomics and Salivaomics Applications
5.1. Infectious Diseases
5.2. Influenza Infection and Possible Antiviral Applications
5.3. Cancer and Degenerative Diseases
5.4. Respiratory Disorders
6. Future Direction and Conclusions
Acknowledgments
Conflicts of Interest
Abbreviations
| ATP | Adenosine triphosphate |
| CE | Capillary electrophoresis |
| CIR | Compensated cirrhosis |
| CKD | Chronic kidney disease |
| CLD | Chronic liver disease |
| COPD | Chronic obstructive pulmonary disease |
| CT scan | Computed tomography scan |
| CV | Coefficient of variability |
| D2O | Deuterium oxide |
| EBC | Exhaled breath condensate |
| EBV | Exhaled breath vapor |
| EEI | Extractive electrospray ionization |
| EI | Electron impact |
| ELISA | Enzyme-linked immunosorbent assay |
| ENO | Exhaled nitric acid |
| FID | free induction decay |
| fMRI | Functional magnetic resonance imaging |
| GC | Gas chromatography |
| HCA | Hierarchical clustering algorithm |
| IBS | Irritable bowel syndrome |
| IR | Infrared |
| LC | Liquid chromatography |
| MS | Mass spectrometry |
| MS | Mass spectrometry |
| NMR | Nuclear magnetic resonance spectroscopy |
| OPLS-DA | Orthogonal partial least squares-discriminate analysis |
| PAH | Pulmonary arterial hypertension |
| PCA | Principle component analysis |
| PLS | Partial least squares |
| PLS-DA | Partial least squares-discriminate analysis |
| PTR | Proton-transfer-reaction |
| SESI | Secondary electrospray Ionization |
| SIFT | Selected ion flow tube |
| SPME | Solid phase microextraction |
| TCA | Tricarboxylic acid |
| TOF | Time-of-flight |
| UHPLC | Ultra-high performance liquid chromatography |
| UV | Ultraviolet |
| UV–vis | Ultraviolet–visible spectroscopy |
| VOCs | Volatile organic compounds |
References
- Bujak, R.; Struck-Lewicka, W.; Markuszewski, M.J.; Kaliszan, R. Metabolomics for laboratory diagnostics. J. Pharm. Biomed. Anal. 2015, 113, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Beale, D.J.; Karpe, A.V.; Ahmed, W. Beyond Metabolomics: A Review of Multi-Omics-Based Approaches. In Microbial Metabolomics: Applications in Clinical, Environmental, and Industrial Microbiology; Beale, D., Kouremenos, K., Palombo, E., Eds.; Springer: Cham, Switzerland, 2016; pp. 295–319. [Google Scholar]
- Snowden, S.; Dahlén, S.-E.; Wheelock, C.E. Application of metabolomics approaches to the study of respiratory diseases. Bioanalysis 2012, 4, 2265–2290. [Google Scholar] [CrossRef] [PubMed]
- Emwas, A.-H.; Luchinat, C.; Turano, P.; Tenori, L.; Roy, R.; Salek, R.M.; Ryan, D.; Merzaban, J.S.; Kaddurah-Daouk, R.; Zeri, A.C.; et al. Standardizing the experimental conditions for using urine in NMR-based metabolomic studies with a particular focus on diagnostic studies: A review. Metabolomics 2015, 11, 872–894. [Google Scholar] [CrossRef] [PubMed]
- Khamis, M.M.; DAdamko, J.; El-Aneed, A. Mass spectrometric based approaches in urine metabolomics and biomarker discovery. Mass Spectrom. Rev. 2015. [Google Scholar] [CrossRef] [PubMed]
- Baraldi, E.; Carraro, S.; Giordano, G.; Reniero, F.; Perilongo, G.; Zacchello, F. Metabolomics: Moving towards personalized medicine. Ital. J. Pediatr. 2009, 35, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Dame, Z.T.; Aziat, F.; Mandal, R.; Krishnamurthy, R.; Bouatra, S.; Borzouie, S.; Guo, A.C.; Sajed, T.; Deng, L.; Lin, H.; et al. The human saliva metabolome. Metabolomics 2015, 11, 1864–1883. [Google Scholar] [CrossRef]
- Yoshizawa, J.M.; Schafer, C.A.; Schafer, J.J.; Farrell, J.J.; Paster, B.J.; Wong, D.T.W. Salivary Biomarkers: Toward Future Clinical and Diagnostic Utilities. Clin. Microbiol. Rev. 2013, 26, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Sun, H.; Wang, X. Saliva metabolomics opens door to biomarker discovery, disease diagnosis, and treatment. Appl. Biochem. Biotechnol. 2012, 168, 1718–1727. [Google Scholar] [CrossRef] [PubMed]
- Pereira, J.; Porto-Figueira, P.; Cavaco, C.; Taunk, K.; Rapole, S.; Dhakne, R.; Nagarajaram, H.; Câmara, J.S. Breath Analysis as a Potential and Non-Invasive Frontier in Disease Diagnosis: An Overview. Metabolites 2015, 5, 3–55. [Google Scholar] [CrossRef] [PubMed]
- Barnes, V.M.; Kennedy, A.D.; Panagakos, F.; Devizio, W.; Trivedi, H.M.; Jönsson, T.; Guo, L.; Cervi, S.; Scannapieco, F.A. Global Metabolomic Analysis of Human Saliva and Plasma from Healthy and Diabetic Subjects, with and without Periodontal Disease. PLoS ONE 2014, 9, e105181. [Google Scholar] [CrossRef] [PubMed]
- Boulpaep, E.L.; Boron, W.F.; Caplan, M.J.; Cantley, L.; Igarashi, P.; Aronson, P.S.; Moczydlowski, E.G. Medical Physiology a Cellular and Molecular Approach. Signal Transduct. 2009, 48, 27. [Google Scholar]
- Humphrey, S.P.; Williamson, R.T. A review of saliva: Normal composition, flow, and function. J. Prosthet. Dent. 2001, 85, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Hibel, L.C.; Granger, D.A.; Cicchetti, D.; Rogosch, F. Salivary biomarker levels and diurnal variation: Associations with medications prescribed to control children’s problem behavior. Child Dev. 2007, 78, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Veerman, E.; Keybus, P.V.D.; Vissink, A.; Amerongen, A. Human glandular salivas: Their separate collection and analysis. Eur. J. Oral Sci. 1996, 104, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Kivlighan, K.T.; Granger, D.A.; Schwartz, E.B. Blood contamination and the measurement of salivary progesterone and estradiol. Horm. Behav. 2005, 47, 367–370. [Google Scholar] [CrossRef] [PubMed]
- Kivlighan, K.T.; Granger, D.A.; Schwartz, E.B.; Nelson, V.; Curran, M.; Shirtcliff, E.A. Quantifying blood leakage into the oral mucosa and its effects on the measurement of cortisol, dehydroepiandrosterone, and testosterone in saliva. Horm. Behav. 2004, 46, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Granger, D.A.; Kivlighan, K.T.; Fortunato, C.; Harmon, A.G.; Hibel, L.C.; Schwartz, E.B.; Whembolua, G.-L. Integration of salivary biomarkers into developmental and behaviorally-oriented research: Problems and solutions for collecting specimens. Physiol. Behav. 2007, 92, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Klein, L.C.; Bennett, J.M.; Whetzel, C.A.; Granger, D.A.; Ritter, F.E. Caffeine and stress alter salivary α-amylase activity in young men. Hum. Psychopharmacol. Clin. Exp. 2010, 25, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Whembolua, G.-L.S.; Granger, D.A.; Singer, S.; Kivlighan, K.T.; Marguin, J.A. Bacteria in the oral mucosa and its effects on the measurement of cortisol, dehydroepiandrosterone, and testosterone in saliva. Horm. Behav. 2006, 49, 478–483. [Google Scholar] [CrossRef] [PubMed]
- Granger, D.A.; Hibel, L.C.; Fortunato, C.K.; Kapelewski, C.H. Medication effects on salivary cortisol: Tactics and strategy to minimize impact in behavioral and developmental science. Psychoneuroendocrinology 2009, 34, 1437–1448. [Google Scholar] [CrossRef] [PubMed]
- Berna, A.Z.; McCarthy, J.S.; Wang, R.X.; Saliba, K.J.; Bravo, F.G.; Cassells, J.; Padovan, B.; Trowell, S.C. Analysis of Breath Specimens for Biomarkers of Plasmodium falciparum Infection. J. Infect. Dis. 2015, 212, 1120–1128. [Google Scholar] [CrossRef] [PubMed]
- Buszewski, B.; Kesy, M.; Ligor, T.; Amann, A. Human exhaled air analytics: Biomarkers of diseases. Biomed. Chromatogr. 2008, 21, 553–566. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.Q.; Duan, Y.X. Breath analysis: Potential for clinical diagnosis and exposure assessment. Clin. Chem. 2006, 52, 800–811. [Google Scholar] [CrossRef] [PubMed]
- Pieil, J.D.; Stiegel, M.A.; Risby, T.H. Clinical breath analysis: Discriminating between human endogenous compounds and exogenous (environmental) chemical confounders. J. Breath Res. 2013, 7, 017107. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, S.; Sugimoto, M.; Kitabatake, K.; Sugano, A.; Nakamura, M.; Kaneko, M.; Ota, S.; Hiwatari, K.; Enomoto, A.; Soga, T.; et al. Identification of salivary metabolomic biomarkers for oral cancer screening. Sci. Rep. 2016, 6, 31520. [Google Scholar] [CrossRef] [PubMed]
- Halama, A.; Kulinski, M.; Kader, S.A.; Satheesh, N.J.; Abou-Samra, A.B.; Suhre, K.; Mohammad, R.M. Measurement of 1,5-anhydroglucitol in blood and saliva: From non-targeted metabolomics to biochemical assay. J. Transl. Med. 2016, 14, 140. [Google Scholar] [CrossRef] [PubMed]
- Tzoulaki, I.; Ebbels, T.M.D.; Valdes, A.; Elliott, P.; Ioannidis, J.P.A. Design and Analysis of Metabolomics Studies in Epidemiological Research: A Primer on-Omic Technologies. Am. J. Epidemiol. 2014, 180, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Ioannidis, J.P.A. Why Most Published Research Findings Are False. PLoS Med. 2005, 2, e124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Button, K.S.; Ioannidis, J.P.A.; Mokrysz, C.; Nosek, B.A.; Flint, J.; Robinson, E.S.J.; Munafo, M.R. Power failure: Why small sample size undermines the reliability of neuroscience. Nat. Rev. Neurosci. 2013, 14, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Mandel, I.D. The Functions of Saliva. J. Dent. Res. 1987, 66, 623–627. [Google Scholar] [CrossRef] [PubMed]
- Daniel, D.A.P.; Thangavel, K. Breathomics for Gastric Cancer Classification Using Back-propagation Neural Network. J. Med. Signals Sens. 2016, 6, 172–182. [Google Scholar] [PubMed]
- Van der Schee, M.P.; Paff, T.; Brinkman, P.; van Aalderen, W.M.; Haarman, E.G.; Sterk, P.J. Breathomics in lung disease. Chest 2015, 147, 224–231. [Google Scholar] [CrossRef] [PubMed]
- May, P.R.A.; van Putten, T.; Jenden, D.J.; Cho, A.K. Test dose response in schizophrenia: Chlorpromazine blood and saliva levels. Arch. Gen. Psychiatry 1978, 35, 1091–1097. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Wong, D.T.; Hirayama, A.; Soga, T.; Tomita, M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics 2010, 6, 78–95. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; St. John, M.A.; Zhou, X. Salivary transcriptome diagnostics for oral cancer detection. Clin. Cancer Res. 2004, 10, 8442–8450. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Gao, P.; Wang, X.; Duan, Y. The early diagnosis and monitoring of squamous cell carcinoma via saliva metabolomics. Sci. Rep. 2014, 4, 6802. [Google Scholar] [CrossRef] [PubMed]
- Pernot, E.; Cardis, E.; Badie, C. Usefulness of Saliva Samples for Biomarker Studies in Radiation Research. Cancer Epidemiol. Biomark. Prev. 2014, 23, 2673–2680. [Google Scholar] [CrossRef] [PubMed]
- Salimetrics, L.; SalivaBio, L. Saliva Collection and Handling Advice. Available online: www.salimetrics.com (accessed on 12 October 2016).
- Dreschel, N.A.; Granger, D.A. Advancing the social neuroscience of human-animal interaction: The role of salivary bioscience. In The Social Neuroscience of Human-Animal Interaction; Freund, L.S., McCune, S., Esposito, L., Gee, N.R., McCardle, P., Eds.; American Psychological Association: Washington, DC, USA, 2016; pp. 195–216. [Google Scholar]
- Sugimoto, M.; Saruta, J.; Matsuki, C.; To, M.; Onuma, H.; Kaneko, M.; Soga, T.; Tomita, M.; Tsukinoki, K. Physiological and environmental parameters associated with mass spectrometry-based salivary metabolomic profiles. Metabolomics 2013, 9, 454–463. [Google Scholar] [CrossRef]
- Goodman, A.M.; Wheelock, M.D.; Harnett, N.G.; Mrug, S.; Granger, D.A.; Knight, D.C. The hippocampal response to psychosocial stress varies with salivary uric acid level. Neuroscience 2016, 339, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Malkar, A.; Devenport, N.A.; Martin, H.J.; Patel, P.; Turner, M.A.; Watson, P.; Maughan, R.J.; Reid, H.J.; Sharp, B.L.; Thomas, C.L.P.; et al. Metabolic profiling of human saliva before and after induced physiological stress by ultra-high performance liquid chromatography—Ion mobility—Mass spectrometry. Metabolomics 2013, 9, 1192–1201. [Google Scholar] [CrossRef] [Green Version]
- Malkar, A.; Wilson, E.; Harrrison, T.; Shaw, D.; Creaser, C. Untargeted metabolic profiling of saliva by liquid chromatography-mass spectrometry for the identification of potential diagnostic biomarkers of asthma. Anal. Methods 2016, 8, 5407–5413. [Google Scholar] [CrossRef] [Green Version]
- Fantuzzi, G.; Righi, E.; Predieri, G.; Ceppelli, G.; Gobba, F.; Aggazzotti, G. Occupational exposure to trihalomethanes in indoor swimming pools. Sci. Total Environ. Health 2001, 264, 257–265. [Google Scholar] [CrossRef]
- Dyne, D.; Cocker, J.; Wilson, H. A novel device for capturing breath samples for solvent analysis. Sci. Total Environ. Health 1997, 199, 83–89. [Google Scholar] [CrossRef]
- Kwak, J.; Fan, M.; Harshman, S.; Garrison, C.; Dershem, V.; Phillips, J.; Grigsby, C.; Ott, D. Evaluation of Bio-VOC Sampler for Analysis of Volatile Organic Compounds in Exhaled Breath. Metabolites 2014, 4, 879. [Google Scholar] [CrossRef] [PubMed]
- Carraro, S.; Rezzi, S.; Reniero, F.; Héberger, K.; Giordano, G.; Zanconato, S.; Guillou, C.; Baraldi, E. Metabolomics applied to exhaled breath condensate in childhood asthma. Am. J. Respir. Crit. Care Med. 2007, 175, 986–990. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, B.; Marsden, P.; Smith, J.A.; Custovic, A.; Nilsson, M.; Fowler, S.J. Breath metabolomic profiling by nuclear magnetic resonance spectroscopy in asthma. Allergy 2013, 68, 1050–1056. [Google Scholar] [CrossRef] [PubMed]
- Nobakht, M.G.B.F.; Aliannejad, R.; Rezaei-Tavirani, M.; Taheri, S.; Oskouie, A.A. The metabolomics of airway diseases, including COPD, asthma and cystic fibrosis. Biomarkers 2015, 20, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.N.; Farquar, G.R.; Jones, A.D.; Frank, M. Human breath analysis: Methods for sample collection and reduction of localized background effects. Anal. Bioanal. Chem. 2010, 396, 739–750. [Google Scholar] [CrossRef] [PubMed]
- Santini, G.; Mores, N.; Penas, A.; Capuano, R.; Mondino, C.; Trové, A.; Macagno, F.; Zini, G.; Cattani, P.; Martinelli, E.; et al. Electronic Nose and Exhaled Breath NMR-based Metabolomics Applications in Airways Disease. Curr. Top. Med. Chem. 2016, 16, 1610–1630. [Google Scholar] [CrossRef] [PubMed]
- Baranska, A.; Mujagic, Z.; Smolinska, A.; Dallinga, J.; Jonkers, D.; Tigchelaar, E.; Dekens, J.; Zhernakova, A.; Ludwig, T.; Masclee, A. Volatile organic compounds in breath as markers for irritable bowel syndrome: A metabolomic approach. Aliment. Pharmacol. Ther. 2016, 44, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.; Penghui, L.; Yunfeng, C.; Wei, Z.; Haidong, W.; Jiao, L.; Jianhua, D.; Huanwen, C. Detection of creatinine in exhaled breath of humans with chronic kidney disease by extractive electrospray ionization mass spectrometry. J. Breath Res. 2016, 10, 016008. [Google Scholar]
- Raymer, J.H.; Thomas, K.W.; Cooper, S.D.; Whitaker, D.A.; Pellizzari, E.D. A device for sampling of human alveolar breath for the measurement of expired volatile organic compounds. J. Anal. Toxicol. 1990, 14, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Perets, T.T.; Shporn, E.; Boltin, D.; Dickman, R.; Niv, Y. Stability of 13C-Urea Breath Test Samples over Time in the Diagnosis of Helicobacter pylori. J. Clin. Lab. Anal. 2016, 30, 231–234. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Paul, T.C.L. How long may a breath sample be stored for at −80 degrees C? A study of the stability of volatile organic compounds trapped onto a mixed Tenax:Carbograph trap adsorbent bed from exhaled breath. J. Breath Res. 2016, 10, 026011. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.; Herrera, J.; Krishnan, S.; Zain, M.; Greenberg, J.; Cataneo, R.N. Variation in volatile organic compounds in the breath of normal humans. J. Chromatogr. B Biomed. Sci. Appl. 1999, 729, 75–88. [Google Scholar] [CrossRef]
- Out, D.; Granger, D.A.; Sephton, S.E.; Segerstrom, S.C. Disentangling sources of individual differences in diurnal salivary α-amylase: Reliability, stability and sensitivity to context. Psychoneuroendocrinology 2013, 38, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Granger, D.A.; Cicchetti, D.; Rogosch, F.A.; Hibel, L.C.; Teisl, M.; Flores, E. Blood contamination in children’s saliva: Prevalence, stability, and impact on the measurement of salivary cortisol, testosterone, and dehydroepiandrosterone. Psychoneuroendocrinology 2007, 32, 724–733. [Google Scholar] [CrossRef] [PubMed]
- Figueira, J.; Jonsson, P.; Adolfsson, A.N.; Adolfsson, R.; Nyberg, L.; Ohman, A. NMR analysis of the human saliva metabolome distinguishes dementia patients from matched controls. Mol. BioSyst. 2016, 12, 2562–2571. [Google Scholar] [CrossRef] [PubMed]
- Oliver, S.G.; Winson, M.K.; Kell, D.B.; Baganz, F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998, 16, 373–378. [Google Scholar] [CrossRef]
- Tweeddale, H.; Notley-McRobb, L.; Ferenci, T. Effect of slow growth on metabolism of Escherichia coli, as revealed by global metabolite pool (“metabolome”) analysis. J. Bacteriol. 1998, 180, 5109–5116. [Google Scholar] [PubMed]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. ‘Metabonomics’: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Keeler, J. Understanding NMR Spectroscopy, 1st ed.; John Wiley and Sons: Chichester, UK, 2005. [Google Scholar]
- Hanson, L.G. Is quantum mechanics necessary for understanding magnetic resonance? Concepts Magn. Reson. A 2008, 32, 329–340. [Google Scholar] [CrossRef]
- Sofia, M.; Maniscalco, M.; de Laurentiis, G.; Paris, D.; Melck, D.; Motta, A. Exploring Airway Diseases by NMR-Based Metabonomics: A Review of Application to Exhaled Breath Condensate. J. Biomed. Biotechnol. 2011, 2011, 403260. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo-García, J.L.; Peces-Barba, G.; Heili, S.; Diaz, R.; Want, E.; Ruiz-Cabello, J. Is NMR-based metabolomic analysis of exhaled breath condensate accurate? Eur. Respir. J. 2011, 37, 468. [Google Scholar] [CrossRef] [PubMed]
- Motta, A.; Paris, D.; Melck, D.; de Laurentiis, G.; Maniscalco, M.; Sofia, M.; Montuschi, P. Nuclear magnetic resonance-based metabolomics of exhaled breath condensate: Methodological aspects. Eur. Respir. J. 2012, 39, 498. [Google Scholar] [CrossRef] [PubMed]
- Rosias, P.P.; Robroeks, C.M.; Kester, A.; den Hartog, G.J.; Wodzig, W.K.; Rijkers, G.T.; Zimmermann, L.J.; van Schayck, C.P.; Jöbsis, Q.; Dompeling, E. Biomarker reproducibility in exhaled breath condensate collected with different condensers. Eur. Respir. J. 2008, 31, 934. [Google Scholar] [CrossRef] [PubMed]
- Koczulla, R.; Dragonieri, S.; Schot, R.; Bals, R.; Gauw, S.A.; Vogelmeier, C.; Rabe, K.F.; Sterk, P.J.; Hiemstra, P.S. Comparison of exhaled breath condensate pH using two commercially available devices in healthy controls, asthma and COPD patients. Respir. Res. 2009, 10, 78. [Google Scholar] [CrossRef] [PubMed]
- Van Mastrigt, E.; de Jongste, J.C.; Pijnenburg, M.W. The analysis of volatile organic compounds in exhaled breath and biomarkers in exhaled breath condensate in children—Clinical tools or scientific toys? Clin. Exp. Allergy 2015, 45, 1170–1188. [Google Scholar] [CrossRef] [PubMed]
- Jones, O.A.H.; Cheung, V.L. An introduction to metabolomics and its potential application in veterinary science. Comp. Med. 2007, 57, 436–442. [Google Scholar] [PubMed]
- Pauling, L.; Robinson, A.B.; Teranishi, R.; Cary, P. Quantitative Analysis of Urine Vapor and Breath by Gas-Liquid Partition Chromatography. Proc. Natl. Acad. Sci. USA 1971, 68, 2374–2376. [Google Scholar] [CrossRef] [PubMed]
- Spinhirne, J.P.; Koziel, J.A.; Chirase, N.K. Sampling and analysis of volatile organic compounds in bovine breath by solid-phase microextraction and gas chromatography-mass spectrometry. J. Chromatogr. A 2004, 1025, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Buszewski, B.; Ulanowska, A.; Ligor, T.; Denderz, N.; Amann, A. Analysis of exhaled breath from smokers, passive smokers and non-smokers by solid-phase microextraction gas chromatography/mass spectrometry. Biomed. Chromatogr. 2009, 23, 551–556. [Google Scholar] [CrossRef] [PubMed]
- Sweetlove, L.J.; Last, R.L.; Fernie, A.R. Predictive Metabolic Engineering: A Goal for Systems Biology. Plant Physiol. 2003, 132, 420–425. [Google Scholar] [CrossRef] [PubMed]
- Fowler, S.J.; Basanta-Sanchez, M.; Xu, Y.; Goodacre, R.; Dark, P.M. Surveillance for lower airway pathogens in mechanically ventilated patients by metabolomic analysis of exhaled breath: A case-control study. Thorax 2015, 70, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Masuo, Y.; Imai, T.; Shibato, J.; Hirano, M.; Jones, O.A.H.; Maguire, M.L.; Satoh, K.; Kikuchi, S.; Rakwal, R. Omic analyses unravels global molecular changes in the brain and liver of a rat model for chronic sake (Japanese alcoholic beverage) intake. Electrophoresis 2009, 30, 1259–1275. [Google Scholar] [CrossRef] [PubMed]
- Kubáň, P.; Kobrin, E.-G.; Kaljurand, M. Capillary electrophoresis—A new tool for ionic analysis of exhaled breath condensate. J. Chromatogr. A 2012, 1267, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Kühnle, M.; Kreidler, D.; Holtin, K.; Czesla, H.; Schuler, P.; Schurig, V.; Albert, K. Online coupling of enantioselective capillary gas chromatography with proton nuclear magnetic resonance spectroscopy. Chirality 2010, 22, 808–812. [Google Scholar] [CrossRef] [PubMed]
- Kühnle, M.; Kreidler, D.; Holtin, K.; Czesla, H.; Schuler, P.; Schaal, W.; Schurig, V.; Albert, K. Online Coupling of Gas Chromatography to Nuclear Magnetic Resonance Spectroscopy: Method for the Analysis of Volatile Stereoisomers. Anal. Chem. 2008, 80, 5481–5486. [Google Scholar] [CrossRef] [PubMed]
- Kim, L.; Mitrevski, B.; Tuck, K.L.; Marriott, P.J. Quantitative preparative gas chromatography of caffeine with nuclear magnetic resonance spectroscopy. J. Sep. Sci. 2013, 36, 1774–1780. [Google Scholar] [CrossRef] [PubMed]
- Park, H.E.; Yang, S.O.; Hyun, S.H.; Park, S.J.; Choi, H.K.; Marriott, P.J. Simple preparative gas chromatographic method for isolation of menthol and menthone from peppermint oil, with quantitative GC-MS and 1H-NMR assay. J. Sep. Sci. 2012, 35, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Dias, D.; Urban, S. Phytochemical analysis of the Southern Australian marine alga, Plocamium mertensii using HPLC-NMR. Phytochem. Anal. 2008, 19, 453–470. [Google Scholar] [CrossRef] [PubMed]
- Urban, S.; Dias, D.A. NMR Spectroscopy: Structure Elucidation of Cycloelatanene A: A Natural Product Case Study. In Metabolomics Tools for Natural Product Discovery: Methods and Protocols; Roessner, U., Dias, A.D., Eds.; Humana Press: Totowa, NJ, USA, 2013; pp. 99–116. [Google Scholar]
- Lamote, K.; Nackaerts, K.; van Meerbeeck, J.P. Strengths, weaknesses, and opportunities of diagnostic breathomics in pleural mesothelioma-A hypothesis. Cancer Epidemiol. Biomark. Prev. 2014, 23, 898–908. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Bean, H.D.; Wargo, M.J.; Leclair, L.W.; Hill, J.E. Detecting bacterial lung infections: In vivo evaluation of in vitro volatile fingerprints. J. Breath Res. 2013, 7, 016003. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Duan, J.; Duan, Y. Recent developments of proton-transfer reaction mass spectrometry (PTR-MS) and its applications in medical research. Mass Spectrom. Rev. 2013, 32, 143–165. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Huang, J.; Abbassi-Ghadi, N.; Španěl, P.; Smith, D.; Hanna, G.B. Selected Ion Flow Tube Mass Spectrometry Analysis of Exhaled Breath for Volatile Organic Compound Profiling of Esophago-Gastric Cancer. Anal. Chem. 2013, 85, 6121–6128. [Google Scholar] [CrossRef] [PubMed]
- Lanucara, F.; Holman, S.W.; Gray, C.J.; Eyers, C.E. The power of ion mobility-mass spectrometry for structural characterization and the study of conformational dynamics. Nat. Chem. 2014, 6, 281–294. [Google Scholar] [CrossRef] [PubMed]
- Halbfeld, C.; Ebert, B.; Blank, L. Multi-Capillary Column-Ion Mobility Spectrometry of Volatile Metabolites Emitted by Saccharomyces Cerevisiae. Metabolites 2014, 4, 751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Handa, H.; Usuba, A.; Maddula, S.; Baumbach, J.I.; Mineshita, M.; Miyazawa, T. Exhaled Breath Analysis for Lung Cancer Detection Using Ion Mobility Spectrometry. PLoS ONE 2014, 9, e114555. [Google Scholar] [CrossRef] [PubMed]
- Miekisch, W.; Schubert, J.K. From highly sophisticated analytical techniques to life-saving diagnostics: Technical developments in breath analysis. TrAC Trends Anal. Chem. 2006, 25, 665–673. [Google Scholar] [CrossRef]
- Wang, C.; Sahay, P. Breath Analysis Using Laser Spectroscopic Techniques: Breath Biomarkers, Spectral Fingerprints, and Detection Limits. Sensors 2009, 9, 8230. [Google Scholar] [CrossRef] [PubMed]
- Chow, K.K.; Short, M.; Zeng, H. A comparison of spectroscopic techniques for human breath analysis. Biomed. Spectrosc. Imaging 2012, 1, 339–353. [Google Scholar]
- Vries, R.D.; Brinkman, P.; Schee, M.P.V.D.; Fens, N.; Dijkers, E.; Bootsma, S.K.; Jongh, F.H.C.D.; Sterk, P.J. Integration of electronic nose technology with spirometry: Validation of a new approach for exhaled breath analysis. J. Breath Res. 2015, 9, 046001. [Google Scholar] [CrossRef] [PubMed]
- Montuschi, P.; Mores, N.; Trové, A.; Mondino, C.; Barnes, P.J. The Electronic Nose in Respiratory Medicine. Respiration 2013, 85, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Saccenti, E.; Hoefsloot, H.C.J.; Smilde, A.K.; Westerhuis, J.A.; Hendriks, M.M.W.B. Reflections on univariate and multivariate analysis of metabolomics data. Metabolomics 2013, 10, 361–374. [Google Scholar] [CrossRef]
- Maiga, M.; Choi, S.W.; Atudorei, V.; Maiga, M.C.; Sharp, Z.D.; Bishai, W.R.; Timmins, G.S. In vitro and In vivo studies of a rapid and selective breath test for tuberculosis based upon mycobacterial CO dehydrogenase. mBio 2014, 5, e00990-14. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Jimenez-Diaz, J.; Bean, H.D.; Daphtary, N.A.; Aliyeva, M.I.; Lundblad, L.K.; Hill, J.E. Robust detection of P. aeruginosa and S. aureus acute lung infections by secondary electrospray ionization-mass spectrometry (SESI-MS) breathprinting: From initial infection to clearance. J. Breath Res. 2013, 7, 037106. [Google Scholar] [CrossRef] [PubMed]
- Bean, H.D.; Jiménez-Díaz, J.; Zhu, J.; Hill, J.E. Breathprints of model murine bacterial lung infections are linked with immune response. Eur. Respir. J. 2015, 45, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Lowen, A.C.; Mubareka, S.; Tumpey, T.M.; Garcia-Sastre, A.; Palese, P. The guinea pig as a transmission model for human influenza viruses. Proc. Natl. Acad. Sci. USA 2006, 103, 9988–9992. [Google Scholar] [CrossRef] [PubMed]
- Ritter, J.B.; Wahl, A.S.; Freund, S.; Genzel, Y.; Reichl, U. Metabolic effects of influenza virus infection in cultured animal cells: Intra- and extracellular metabolite profiling. BMC Syst. Biol. 2010, 4, 61. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Liu, N.; Yang, Z.; Song, W.; Wang, P.; Chen, H.; Lucio, M.; Schmitt-Kopplin, P.; Chen, G.; Cai, Z. GC/MS-based metabolomics reveals fatty acid biosynthesis and cholesterol metabolism in cell lines infected with influenza A virus. Talanta 2010, 83, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Fan, J.; Li, Y.; Shi, X.; Ju, D.; Yan, Q.; Yan, X.; Han, L.; Zhu, H. Modified Jiu Wei Qiang Huo decoction improves dysfunctional metabolomics in influenza A pneumonia-infected mice. Biomed. Chromatogr. 2014, 28, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.H.; Golden, J.E.; Adcock, R.S.; Schroeder, C.E.; Chu, Y.K.; Sotsky, J.B.; Cramer, D.E.; Chilton, P.M.; Song, C.; Anantpadma, M.; et al. Discovery of a Broad-Spectrum Antiviral Compound That Inhibits Pyrimidine Biosynthesis and Establishes a Type 1 Interferon-Independent Antiviral State. Antimicrob. Agents Chemother. 2016, 60, 4552–4562. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Gaelings, L.; Soderholm, S.; Belanov, S.; Nandania, J.; Nyman, T.A.; Matikainen, S.; Anders, S.; Velagapudi, V.; Kainov, D.E. JNJ872 inhibits influenza A virus replication without altering cellular antiviral responses. Antivir. Res. 2016, 133, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Rabinowitz, J.D.; Purdy, J.G.; Vastag, L.; Shenk, T.; Koyuncu, E. Metabolomics in drug target discovery. Cold Spring Harb. Symp. Quant. Biol. 2011, 76, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Aksenov, A.A.; Sandrock, C.E.; Zhao, W.; Sankaran, S.; Schivo, M.; Harper, R.; Cardona, C.J.; Xing, Z.; Davis, C.E. Cellular Scent of Influenza Virus Infection. Chembiochem Eur. J. Chem. Biol. 2014, 15, 1040–1048. [Google Scholar] [CrossRef] [PubMed]
- Berna, A.Z.; McCarthy, J.S.; Trowell, S.C. Malaria detection using breath biomarkers. Med. J. Aust. 2016, 204, 50. [Google Scholar] [CrossRef] [PubMed]
- Jianping, G.; Yingchang, Z.; Yonggang, W.; Feng, W.; Lang, L.; Ping, W.; Yong, Z.; Kejing, Y. Breath analysis for noninvasively differentiating Acinetobacter baumannii ventilator-associated pneumonia from its respiratory tract colonization of ventilated patients. J. Breath Res. 2016, 10, 027102. [Google Scholar]
- Chandler, J.D.; Hu, X.; Ko, E.J.; Park, S.; Lee, Y.T.; Orr, M.L.; Fernandes, J.; Uppal, K.; Kang, S.M.; Jones, D.P.; et al. Metabolic pathways of lung inflammation revealed by high-resolution metabolomics (HRM) of H1N1 influenza virus infection in mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2016, 311, R906–R916. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Zheng, D.; Lee, Y.H.; Chan, T.K.; Kumar, Y.; Ho, W.E.; Chen, J.Z.; Tannenbaum, S.R.; Ong, C.N. Metabolomics Investigation Reveals Metabolite Mediators Associated with Acute Lung Injury and Repair in a Murine Model of Influenza Pneumonia. Sci. Rep. 2016, 6, 26076. [Google Scholar] [CrossRef] [PubMed]
- Jose, L.I.-G.; Jesús, R.-C.; Pablo, C.; Pilar, F.-S.; Andrés, E.; José, A.L. Metabolomic Analysis as a Diagnostic Tool for Acute Respiratory Distress Syndrome Caused by H1N1 Influenza Infection in Humans. In A25. Predicting Development and Outcomes in Acute Lung Injury; American Thoracic Society: New York, NY, USA, 2012; p. A1149. [Google Scholar]
- Phillips, M.; Cataneo, R.N.; Chaturvedi, A.; Danaher, P.J.; Devadiga, A.; Legendre, D.A.; Nail, K.L.; Schmitt, P.; Wai, J. Effect of influenza vaccination on oxidative stress products in breath. J. Breath Res. 2010, 4, 026001. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.Y.; Hurt, A.C. Using the ferret as an animal model for investigating influenza antiviral effectiveness. Front. Microbiol. 2016, 7, 80. [Google Scholar] [CrossRef] [PubMed]
- Gustin, K.M.; Katz, J.M.; Tumpey, T.M.; Maines, T.R. Comparison of the levels of infectious virus in respirable aerosols exhaled by ferrets infected with influenza viruses exhibiting diverse transmissibility phenotypes. J. Virol. 2013, 87, 7864–7873. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, Y.; Lamirande, E.W.; Subbarao, K. The ferret model for influenza. In Current Protocols in Microbiology; Wiley: Hoboken, NJ, USA, 2009; Chapter 15, Unit 15G.2. [Google Scholar]
- Belser, J.A.; Eckert, A.M.; Tumpey, T.M.; Maines, T.R. Complexities in Ferret Influenza Virus Pathogenesis and Transmission Models. Microbiol. Mol. Biol. Rev. 2016, 80, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, G.; Ichikawa, Y.; Yoshitomi, I.; Umeda, M. Metabolomics of Salivary Biomarkers in Yusho Patients. Fukuoka Igaku Zasshi 2015, 106, 144–148. [Google Scholar] [PubMed]
- Mueller, D.C.; Piller, M.; Niessner, R.; Scherer, M.; Scherer, G. Untargeted Metabolomic Profiling in Saliva of Smokers and Nonsmokers by a Validated GC-TOF-MS Method. J. Proteome Res. 2014, 13, 1602–1613. [Google Scholar] [CrossRef] [PubMed]
- Bach, J.-P.; Gold, M.; Mengel, D.; Hattesohl, A.; Lubbe, D.; Schmid, S.; Tackenberg, B.; Rieke, J.; Maddula, S.; Baumbach, J.I.; et al. Measuring Compounds in Exhaled Air to Detect Alzheimer’s Disease and Parkinson’s Disease. PLoS ONE 2015, 10, e0132227. [Google Scholar] [CrossRef] [PubMed]
- Pijls, K.E.; Smolinska, A.; Jonkers, D.M.A.E.; Dallinga, J.W.; Masclee, A.A.M.; Koek, G.H.; van Schooten, F.-J. A profile of volatile organic compounds in exhaled air as a potential non-invasive biomarker for liver cirrhosis. Sci. Rep. 2016, 6, 19903. [Google Scholar] [CrossRef] [PubMed]
- Horváth, I.; Lázár, Z.; Gyulai, N.; Kollai, M.; Losonczy, G. Exhaled biomarkers in lung cancer. Eur. Respir. J. 2009, 34, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Peralbo-Molina, A.; Calderón-Santiago, M.; Priego-Capote, F.; Jurado-Gámez, B.; Castro, M.D.L.D. Metabolomics analysis of exhaled breath condensate for discrimination between lung cancer patients and risk factor individuals. J. Breath Res. 2016, 10, 016011. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, U.; Naiker, M.; Myers, M. Cell culture metabolomics in the diagnosis of lung cancer—The influence of cell culture conditions. J. Breath Res. 2014, 8, 027109. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Ahmed, S. Emerging field of metabolomics: Big promise for cancer biomarker identification and drug discovery. J. Pharm. Biomed. Anal. 2015, 107, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Institute for Health Metrics and Evaluation, Global Burden of Disease Study. 2014. Available online: http://www.healthdata.org/gbd (accessed on 10 October 2016).
- Dahlin, A.; McGeachie, M.J.; Lasky-Su, J.A. Asthma Metabolomics: The Missing Step for Translating Bench Work into the Clinic. J. Pulm. Respir. Med. 2015, 5, 267. [Google Scholar]
- Luxon, B.A. Metabolomics in asthma. Adv. Exp. Med. Biol. 2014, 795, 207–220. [Google Scholar] [PubMed]
- Comhair, S.A.; McDunn, J.; Bennett, C.; Fettig, J.; Erzurum, S.C.; Kalhan, S.C. Metabolomic Endotype of Asthma. J. Immunol. 2015, 195, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Adamko, D.J.; Sykes, B.D.; Rowe, B.H. The metabolomics of asthma: Novel diagnostic potential. Chest 2012, 141, 1295–1302. [Google Scholar] [CrossRef] [PubMed]
- Motta, A.; Paris, D.; D’Amato, M.; Melck, D.; Calabrese, C.; Vitale, C.; Stanziola, A.A.; Corso, G.; Sofia, M.; Maniscalco, M. NMR metabolomic analysis of exhaled breath condensate of asthmatic patients at two different temperatures. J. Proteome Res. 2014, 13, 6107–6120. [Google Scholar] [CrossRef] [PubMed]
- Guillevin, L.; Armstrong, I.; Aldrighetti, R.; Howard, L.S.; Ryftenius, H.; Fischer, A.; Lombardi, S.; Studer, S.; Ferrari, P. Understanding the impact of pulmonary arterial hypertension on patients’ and carers’ lives. Eur. Respir. Rev. 2013, 22, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Peng, J.; Lu, C.; Hsin, M.; Mura, M.; Wu, L.; Chu, L.; Zamel, R.; Machuca, T.; Waddell, T.; et al. Metabolomic Heterogeneity of Pulmonary Arterial Hypertension. PLoS ONE 2014, 9, e88727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cikach, F.S.; Tonelli, A.R.; Barnes, J.; Paschke, K.; Newman, J.; Grove, D.; Dababneh, L.; Wang, S.; Dweik, R.A. Breath Analysis in Pulmonary Arterial Hypertension. Chest 2014, 145, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Bos, L.D.; Weda, H.; Wang, Y.; Knobel, H.H.; Nijsen, T.M.; Vink, T.J.; Zwinderman, A.H.; Sterk, P.J.; Schultz, M.J. Exhaled breath metabolomics as a noninvasive diagnostic tool for acute respiratory distress syndrome. Eur. Respir. J. 2014, 44, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Stringer, K.A.; McKay, R.T.; Karnovsky, A.; Quémerais, B.; Lacy, P. Metabolomics and Its Application to Acute Lung Diseases. Front. Immunol. 2016, 7, 44. [Google Scholar] [CrossRef] [PubMed]
- Kostikas, K.; Papatheodorou, G.; Psathakis, K.; Panagou, P.; Loukides, S. Oxidative stress in expired breath condensate of patients with COPD. Chest 2003, 124, 1373–1380. [Google Scholar] [CrossRef] [PubMed]
- Muhlebach, M.S.; Sha, W. Lessons learned from metabolomics in cystic fibrosis. Mol. Cell. Pediatr. 2015, 2, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Montuschi, P.; Paris, D.; Melck, D.; Lucidi, V.; Ciabattoni, G.; Raia, V.; Calabrese, C.; Bush, A.; Barnes, P.J.; Motta, A. NMR spectroscopy metabolomic profiling of exhaled breath condensate in patients with stable and unstable cystic fibrosis. Thorax 2012, 67, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Montuschi, P.; Paris, D.; Montella, S.; Melck, D.; Mirra, V.; Santini, G.; Mores, N.; Montemitro, E.; Majo, F.; Lucidi, V.; et al. Nuclear Magnetic Resonance–based Metabolomics Discriminates Primary Ciliary Dyskinesia from Cystic Fibrosis. Am. J. Respir. Crit. Care Med. 2014, 190, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Paris, D.; Maniscalco, M.; Melck, D.; D’Amato, M.; Sorrentino, N.; Zedda, A.; Sofia, M.; Motta, A. Inflammatory metabolites in exhaled breath condensate characterize the obese respiratory phenotype. Metabolomics 2015, 11, 1934–1939. [Google Scholar] [CrossRef]
| Breath Type | Characteristics | ||
|---|---|---|---|
| Collection Device | Advantages | Disadvantages | |
| Initial |
|
|
|
| Modified (alveolar breath) |
|
|
|
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beale, D.J.; Jones, O.A.H.; Karpe, A.V.; Dayalan, S.; Oh, D.Y.; Kouremenos, K.A.; Ahmed, W.; Palombo, E.A. A Review of Analytical Techniques and Their Application in Disease Diagnosis in Breathomics and Salivaomics Research. Int. J. Mol. Sci. 2017, 18, 24. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18010024
Beale DJ, Jones OAH, Karpe AV, Dayalan S, Oh DY, Kouremenos KA, Ahmed W, Palombo EA. A Review of Analytical Techniques and Their Application in Disease Diagnosis in Breathomics and Salivaomics Research. International Journal of Molecular Sciences. 2017; 18(1):24. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18010024
Chicago/Turabian StyleBeale, David J., Oliver A. H. Jones, Avinash V. Karpe, Saravanan Dayalan, Ding Yuan Oh, Konstantinos A. Kouremenos, Warish Ahmed, and Enzo A. Palombo. 2017. "A Review of Analytical Techniques and Their Application in Disease Diagnosis in Breathomics and Salivaomics Research" International Journal of Molecular Sciences 18, no. 1: 24. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18010024