A Comprehensive Survey of the Roles of Highly Disordered Proteins in Type 2 Diabetes
Abstract
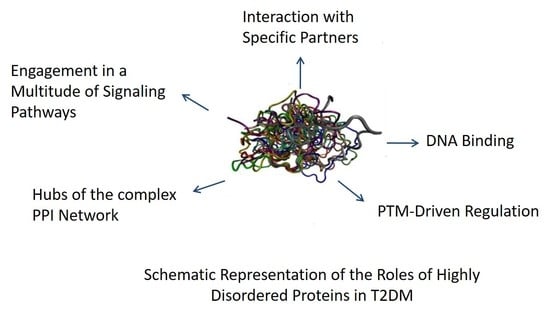
:1. Introduction
2. Results and Discussion
2.1. The Overall Intrinsic Disorder Status of the Type 2 Diabetes Mellitus (T2DM)-Related Proteins
2.2. Musculoaponeurotic Fibrosarcoma (MAF) bZIP Transcription Factor A (MAFA, UniProt ID: Q8NHW3, PONDR® FIT: 73.2%)
2.2.1. Domain Structure of MAF Proteins
2.2.2. Intrinsic Disorder Status of Human MAFA Protein
2.2.3. Functionality of Intrinsic Disorder in Human MAFA Protein
2.3. Insulin Receptor Substrates, IRS1 (UniProt: P35568, PONDR® FIT: 70.7%), IRS2 (UniProt: Q9Y4H2, PONDR® FIT: 75.6%), and IRS4 (UniProt: O14654, PONDR® FIT: 64.4%)
2.3.1. Domain Structure of the Insulin Receptor Substrates
2.3.2. Order and Intrinsic Disorder in Functionality of Human IRS1, IRS2, and IRS4 Proteins
2.4. Pancreatic and Duodenal Homeobox 1 Protein, PDX1 (UniProt ID: P52945, PONDR® FIT: 60.4%)
2.4.1. Domain Structure of the PDX1 Protein
2.4.2. Prevalence of Functional Intrinsic Disorder in Human PDX1 Protein
2.5. Adiponectin (UniProt ID: Q15848; PONDR® FIT: 38.9%)
2.6. Phosphoinositide 3-Kinase Regulatory Subunits 2 and 5, PIK3R2 (UniProt ID: O00459; PONDR® FIT: 34.1%), and PIK3R5 (UniProt ID: Q8WYR1; PONDR® FIT: 36.7%)
2.6.1. Functionality of Human PI3K Proteins
2.6.2. Structural Organization and Functional Intrinsic Disorder of Human PIK3R2
2.7. Suppressors of Cytokine Signaling, SoCS1 (UniProt ID: O15524, PONDR® FIT: 34.6%) and SoCS3 (UniProt ID: O14543, PONDR® FIT: 32.0%)
2.8. Insulin (UniProt ID: P01308, PONDR® FIT: 16.3%)
2.8.1. Function and Structural Organization of Human Insulin
2.8.2. Prevalence and Functionality of Intrinsic Disorder of Human Insulin
2.9. Insulin Receptor (UniProt ID: P06213, PONDR® FIT: 14.0%)
2.9.1. Functional Roles of Human Insulin Receptor
2.9.2. Structural Organization and Functional Intrinsic Disorder of Human IR
3. Experimental Section
3.1. Dataset
3.2. Computational Analyses of the Amino Acid Sequences of T2DM Biomarkers
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bergman, R.N.; Ader, M.; Huecking, K.; van Citters, G. Accurate assessment of β-cell function: The hyperbolic correction. Diabetes 2002, 51 (Suppl. 1), S212–S220. [Google Scholar] [CrossRef] [PubMed]
- Whiting, D.R.; Guariguata, L.; Weil, C.; Shaw, J. IDF diabetes atlas: Global estimates of the prevalence of diabetes for 2011 and 2030. Diabetes Res. Clin. Pract. 2011, 94, 311–321. [Google Scholar] [CrossRef] [PubMed]
- NCD Risk Factor Collaboration. Worldwide trends in diabetes since 1980: A pooled analysis of 751 population-based studies with 4.4 million participants. Lancet 2016, 387, 1513–1530. [Google Scholar]
- Morandi, A.; Maffeis, C. Predictors of metabolic risk in childhood obesity. Horm. Res. Paediatr. 2014, 82, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Adebayo, O.; Willis, G.C. The changing face of diabetes in America. Emerg. Med. Clin. N. Am. 2014, 32, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Prasad, H.C. Changing epidemiology of metabolic syndrome and type 2 diabetes in chinese youth. Curr. Diabetes Rep. 2014, 14, 447. [Google Scholar] [CrossRef] [PubMed]
- Pinhas-Hamiel, O.; Zeitler, P. The global spread of type 2 diabetes mellitus in children and adolescents. J. Pediatr. 2005, 146, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Huxley, R.; Wu, Y.; Dibley, M.J. Temporal trends in overweight and obesity of children and adolescents from nine provinces in china from 1991–2006. Int. J. Pediatr. Obes. 2010, 5, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.F.; Liang, L.; Gong, C.X.; Xiong, F.; Luo, F.H.; Liu, G.L.; Li, P.; Liu, L.; Xin, Y.; Yao, H.; et al. Status and trends of diabetes in Chinese children: Analysis of data from 14 medical centers. World J. Pediatr. 2013, 9, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Dabelea, D.; DeGroat, J.; Sorrelman, C.; Glass, M.; Percy, C.A.; Avery, C.; Hu, D.; D’Agostino, R.B., Jr.; Beyer, J.; Imperatore, G.; et al. Diabetes in navajo youth: Prevalence, incidence, and clinical characteristics: The search for diabetes in youth study. Diabetes Care 2009, 32 (Suppl. 2), S141–S147. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.L.; Yi, J.P.; Beyer, J.; Mayer-Davis, E.J.; Dolan, L.M.; Dabelea, D.M.; Lawrence, J.M.; Rodriguez, B.L.; Marcovina, S.M.; Waitzfelder, B.E.; et al. Type 1 and type 2 diabetes in Asian and Pacific Islander U.S. Youth. Diabetes Care 2009, 32 (Suppl. 2), S133–S140. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, J.M.; Mayer-Davis, E.J.; Reynolds, K.; Beyer, J.; Pettitt, D.J.; D’Agostino, R.B., Jr.; Marcovina, S.M.; Imperatore, G.; Hamman, R.F.; SEARCH for Diabetes in Youth Study Group. Diabetes in hispanic american youth: Prevalence, incidence, demographics, and clinical characteristics. Diabetes Care 2009, 32 (Suppl. 2), S123–S132. [Google Scholar] [CrossRef] [PubMed]
- Mayer-Davis, E.J.; Beyer, J.; Bell, R.A.; Dabelea, D.; D’Agostino, R., Jr.; Imperatore, G.; Lawrence, J.M.; Liese, A.D.; Liu, L.; Marcovina, S.; et al. Diabetes in african american youth: Prevalence, incidence, and clinical characteristics. Diabetes Care 2009, 32 (Suppl. 2), S112–122. [Google Scholar] [CrossRef] [PubMed]
- Bell, R.A.; Mayer-Davis, E.J.; Beyer, J.W.; D’Agostino, R.B., Jr.; Lawrence, J.M.; Linder, B.; Liu, L.L.; Marcovina, S.M.; Rodriguez, B.L.; Williams, D.; et al. Diabetes in non-hispanic white youth: Prevalence, incidence, and clinical characteristics. Diabetes Care 2009, 32 (Suppl. 2), S102–S111. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, T.A.; Kaneto, H.; Miyatsuka, T.; Yamamoto, T.; Yamamoto, K.; Kato, K.; Shimomura, I.; Stein, R.; Matsuhisa, M. Regulation of mafa expression in pancreatic β-cells in db/db mice with diabetes. Diabetes 2010, 59, 1709–1720. [Google Scholar] [CrossRef] [PubMed]
- Mahadevan, J.; Parazzoli, S.; Oseid, E.; Hertzel, A.V.; Bernlohr, D.A.; Vallerie, S.N.; Liu, C.Q.; Lopez, M.; Harmon, J.S.; Robertson, R.P. Ebselen treatment prevents islet apoptosis, maintains intranuclear Pdx-1 and MafA levels, and preserves β-cell mass and function in ZDF rats. Diabetes 2013, 62, 3582–3588. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Dai, C.; Guo, M.; Taylor, B.; Harmon, J.S.; Sander, M.; Robertson, R.P.; Powers, A.C.; Stein, R. Inactivation of specific β cell transcription factors in type 2 diabetes. J. Clin. Investig. 2013, 123, 3305–3316. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Furumichi, M.; Tanabe, M.; Hirakawa, M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010, 38, D355–D360. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Oldfield, C.J.; Dunker, A.K. Intrinsically disordered proteins in human diseases: Introducing the D2 concept. Annu. Rev. Biophys. 2008, 37, 215–246. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. Pondr-fit: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 2010, 1804, 996–1010. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence complexity of disordered protein. Proteins 2001, 42, 38–48. [Google Scholar] [CrossRef]
- Iakoucheva, L.M.; Radivojac, P.; Brown, C.J.; O’Connor, T.R.; Sikes, J.G.; Obradovic, Z.; Dunker, A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004, 32, 1037–1049. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Vucetic, S.; Radivojac, P.; Brown, C.J.; Dunker, A.K.; Obradovic, Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J. Bioinform. Comput. Biol. 2005, 3, 35–60. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Radivojac, P.; Vucetic, S.; Dunker, A.K.; Obradovic, Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinform. 2006, 7, 208. [Google Scholar] [CrossRef] [PubMed]
- Dosztanyi, Z.; Csizmok, V.; Tompa, P.; Simon, I. Iupred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 2005, 21, 3433–3434. [Google Scholar] [CrossRef] [PubMed]
- Dyson, H.J.; Wright, P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005, 6, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Gsponer, J.; Babu, M.M. The rules of disorder or why disorder rules. Prog. Biophys. Mol. Biol. 2009, 99, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, K.; Mooney, S.M.; Parekh, N.; Getzenberg, R.H.; Kulkarni, P. A majority of the cancer/testis antigens are intrinsically disordered proteins. J. Cell. Biochem. 2011, 112, 3256–3267. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Romero, P.; Rani, M.; Dunker, A.K.; Obradovic, Z. Predicting protein disorder for N-,C-, and internal regions. Genome Inform. Ser. Workshop Genome Inform. 1999, 10, 30–40. [Google Scholar] [PubMed]
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 2000, 41, 415–427. [Google Scholar] [CrossRef]
- Oldfield, C.J.; Cheng, Y.; Cortese, M.S.; Brown, C.J.; Uversky, V.N.; Dunker, A.K. Comparing and combining predictors of mostly disordered proteins. Biochemistry 2005, 44, 1989–2000. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Oldfield, C.J.; Xue, B.; Hsu, W.L.; Meng, J.; Liu, X.; Shen, L.; Romero, P.; Uversky, V.N.; Dunker, A. Improving protein order-disorder classification using charge-hydropathy plots. BMC Bioinform. 2014, 15 (Suppl. 17), S4. [Google Scholar]
- Blank, V.; Andrews, N.C. The maf transcription factors: Regulators of differentiation. Trends Biochem. Sci. 1997, 22, 437–441. [Google Scholar] [CrossRef]
- Kataoka, K.; Han, S.I.; Shioda, S.; Hirai, M.; Nishizawa, M.; Handa, H. MafA is a glucose-regulated and pancreatic β-cell-specific transcriptional activator for the insulin gene. J. Biol. Chem. 2002, 277, 49903–49910. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, T.A.; Artner, I.; Henderson, E.; Means, A.; Sander, M.; Stein, R. The MafA transcription factor appears to be responsible for tissue-specific expression of insulin. Proc. Natl. Acad. Sci. USA 2004, 101, 2930–2933. [Google Scholar] [CrossRef] [PubMed]
- Benkhelifa, S.; Provot, S.; Lecoq, O.; Pouponnot, C.; Calothy, G.; Felder-Schmittbuhl, M.P. MafA, a novel member of the maf proto-oncogene family, displays developmental regulation and mitogenic capacity in avian neuroretina cells. Oncogene 1998, 17, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, K.; Fujiwara, K.T.; Noda, M.; Nishizawa, M. MafB, a new Maf family transcription activator that can associate with Maf and Fos but not with Jun. Mol. Cell. Biol. 1994, 14, 7581–7591. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, K.; Nishizawa, M.; Kawai, S. Structure-function analysis of the Maf oncogene product, a member of the b-Zip protein family. J. Virol. 1993, 67, 2133–2141. [Google Scholar] [PubMed]
- Swaroop, A.; Xu, J.Z.; Pawar, H.; Jackson, A.; Skolnick, C.; Agarwal, N. A conserved retina-specific gene encodes a basic motif/leucine zipper domain. Proc. Natl. Acad. Sci. USA 1992, 89, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Guo, M.; Matsuoka, T.A.; Hagman, D.K.; Parazzoli, S.D.; Poitout, V.; Stein, R. The islet β cell-enriched MafA activator is a key regulator of insulin gene transcription. J. Biol. Chem. 2005, 280, 11887–11894. [Google Scholar] [CrossRef] [PubMed]
- Rocques, N.; Abou Zeid, N.; Sii-Felice, K.; Lecoin, L.; Felder-Schmittbuhl, M.P.; Eychene, A.; Pouponnot, C. GSK-3-mediated phosphorylation enhances Maf-transforming activity. Mol. Cell 2007, 28, 584–597. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, T.A.; Kaneto, H.; Kawashima, S.; Miyatsuka, T.; Tochino, Y.; Yoshikawa, A.; Imagawa, A.; Miyazaki, J.; Gannon, M.; Stein, R.; et al. Preserving MafA expression in diabetic islet β-cells improves glycemic control in vivo. J. Biol. Chem. 2015, 290, 7647–7657. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, H.; Motohashi, H.; Sueno, S.; Kimura, M.; Takagawa, H.; Kanno, Y.; Yamamoto, M.; Tanaka, T. Structural basis of alternative DNA recognition by Maf transcription factors. Mol. Cell. Biol. 2009, 29, 6232–6244. [Google Scholar] [CrossRef] [PubMed]
- Fuxreiter, M.; Simon, I.; Bondos, S. Dynamic protein-DNA recognition: Beyond what can be seen. Trends Biochem. Sci. 2011, 36, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Minezaki, Y.; Homma, K.; Kinjo, A.R.; Nishikawa, K. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcriptional regulation. J. Mol. Biol. 2006, 359, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Perumal, N.B.; Oldfield, C.J.; Su, E.W.; Uversky, V.N.; Dunker, A.K. Intrinsic disorder in transcription factors. Biochemistry 2006, 45, 6873–6888. [Google Scholar] [CrossRef] [PubMed]
- Eychene, A.; Rocques, N.; Pouponnot, C. A new mafia in cancer. Nat. Rev. Cancer 2008, 8, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Obradovic, Z.; Peng, K.; Vucetic, S.; Radivojac, P.; Dunker, A.K. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins 2005, 61 (Suppl. 7), 176–182. [Google Scholar] [CrossRef] [PubMed]
- Oates, M.E.; Romero, P.; Ishida, T.; Ghalwash, M.; Mizianty, M.J.; Xue, B.; Dosztanyi, Z.; Uversky, V.N.; Obradovic, Z.; Kurgan, L.; et al. D2P2: Database of disordered protein predictions. Nucleic Acids Res. 2013, 41, D508–D516. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Kinoshita, K. Prdos: Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007, 35, W460–W464. [Google Scholar] [CrossRef] [PubMed]
- Walsh, I.; Martin, A.J.; Di Domenico, T.; Tosatto, S.C. Espritz: Accurate and fast prediction of protein disorder. Bioinformatics 2012, 28, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Pejaver, V.; Hsu, W.L.; Xin, F.; Dunker, A.K.; Uversky, V.N.; Radivojac, P. The structural and functional signatures of proteins that undergo multiple events of post-translational modification. Protein Sci. 2014, 23, 1077–1093. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.E.; Dyson, H.J. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999, 293, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Y.; Chu, X.; Hagen, S.J.; Han, W.; Wang, E. Multi-scaled explorations of binding-induced folding of intrinsically disordered protein inhibitor IA3 to its target enzyme. PLoS Comput. Biol. 2011, 7, e1001118. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.; Wang, Y.; Gan, L.; Bai, Y.; Han, W.; Wang, E.; Wang, J. Importance of electrostatic interactions in the association of intrinsically disordered histone chaperone Chz1 and histone H2A.Z-H2B. PLoS Comput. Biol. 2012, 8, e1002608. [Google Scholar] [CrossRef] [PubMed]
- Vuzman, D.; Levy, Y. Intrinsically disordered regions as affinity tuners in protein-DNA interactions. Mol. Biosyst. 2012, 8, 47–57. [Google Scholar] [CrossRef] [PubMed]
- De Sancho, D.; Best, R.B. Modulation of an IDP binding mechanism and rates by helix propensity and non-native interactions: Association of HIF1α with CBP. Mol. Biosyst. 2012, 8, 256–267. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.E.; Dyson, H.J. Linking folding and binding. Curr. Opin. Struct. Biol. 2009, 19, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Oldfield, C.J.; Cheng, Y.; Cortese, M.S.; Romero, P.; Uversky, V.N.; Dunker, A.K. Coupled folding and binding with α-helix-forming molecular recognition elements. Biochemistry 2005, 44, 12454–12470. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Oldfield, C.J.; Meng, J.; Romero, P.; Uversky, V.N.; Dunker, A.K. Mining α-helix-forming molecular recognition features with cross species sequence alignments. Biochemistry 2007, 46, 13468–13477. [Google Scholar] [CrossRef] [PubMed]
- Meszaros, B.; Simon, I.; Dosztanyi, Z. Prediction of protein binding regions in disordered proteins. PLoS Comput. Biol. 2009, 5, e1000376. [Google Scholar] [CrossRef] [PubMed]
- Dosztanyi, Z.; Meszaros, B.; Simon, I. Anchor: Web server for predicting protein binding regions in disordered proteins. Bioinformatics 2009, 25, 2745–2746. [Google Scholar] [CrossRef] [PubMed]
- Pang, E.; Hao, Y.; Sun, Y.; Lin, K. Differential variation patterns between hubs and bottlenecks in human protein-protein interaction networks. BMC Evol. Biol. 2016, 16, 260. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.; Mason, S.P.; Barabasi, A.L.; Oltvai, Z.N. Lethality and centrality in protein networks. Nature 2001, 411, 41–42. [Google Scholar] [CrossRef] [PubMed]
- Vallabhajosyula, R.R.; Chakravarti, D.; Lutfeali, S.; Ray, A.; Raval, A. Identifying hubs in protein interaction networks. PLoS ONE 2009, 4, e5344. [Google Scholar] [CrossRef] [PubMed]
- Patil, A.; Kinoshita, K.; Nakamura, H. Domain distribution and intrinsic disorder in hubs in the human protein–Wprotein interaction network. Protein Sci. 2010, 19, 1461–1468. [Google Scholar] [CrossRef] [PubMed]
- Patil, A.; Kinoshita, K.; Nakamura, H. Hub promiscuity in protein-protein interaction networks. Int. J. Mol. Sci. 2010, 11, 1930–1943. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The string database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011, 39, D561–D568. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Lopez, D.; Gutierrez, M.A.; Lopes, K.P.; Prieto, C.; Santamaria, R.; de Las Rivas, J. APID interactomes: Providing proteome-based interactomes with controlled quality for multiple species and derived networks. Nucleic Acids Res. 2016, 44, W529–W535. [Google Scholar] [CrossRef] [PubMed]
- Stelzl, U.; Worm, U.; Lalowski, M.; Haenig, C.; Brembeck, F.H.; Goehler, H.; Stroedicke, M.; Zenkner, M.; Schoenherr, A.; Koeppen, S.; et al. A human protein-protein interaction network: A resource for annotating the proteome. Cell 2005, 122, 957–968. [Google Scholar] [CrossRef] [PubMed]
- Huttlin, E.L.; Bruckner, R.J.; Paulo, J.A.; Cannon, J.R.; Ting, L.; Baltier, K.; Colby, G.; Gebreab, F.; Gygi, M.P.; Parzen, H.; et al. Architecture of the human interactome defines protein communities and disease networks. Nature 2017, 545, 505–509. [Google Scholar] [CrossRef] [PubMed]
- Han, S.I.; Aramata, S.; Yasuda, K.; Kataoka, K. MafA stability in pancreatic β cells is regulated by glucose and is dependent on its constitutive phosphorylation at multiple sites by glycogen synthase kinase 3. Mol. Cell. Biol. 2007, 27, 6593–6605. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Guanga, G.P.; Wan, C.; Rose, R.B. A novel DNA binding mechanism for maf basic region-leucine zipper factors inferred from a MafA-DNA complex structure and binding specificities. Biochemistry 2012, 51, 9706–9717. [Google Scholar] [CrossRef] [PubMed]
- Hang, Y.; Stein, R. MafA and MafB activity in pancreatic β cells. Trends Endocrinol. Metab. 2011, 22, 364–373. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, S.; Annoni, C.; Contini, A.; Clerici, F.; Gelmi, M.L. Expedient chemical synthesis of 75mer DNA binding domain of MafA: An insight on its binding to insulin enhancer. Amino Acids 2012, 43, 1995–2003. [Google Scholar] [CrossRef] [PubMed]
- O’Neil, K.T.; Shuman, J.D.; Ampe, C.; DeGrado, W.F. DNA-induced increase in the α-helical content of C/EBP and GCN4. Biochemistry 1991, 30, 9030–9034. [Google Scholar] [CrossRef] [PubMed]
- Weiss, M.A.; Ellenberger, T.; Wobbe, C.R.; Lee, J.P.; Harrison, S.C.; Struhl, K. Folding transition in the DNA-binding domain of GCN4 on specific binding to DNA. Nature 1990, 347, 575–578. [Google Scholar] [CrossRef] [PubMed]
- Fujii, Y.; Shimizu, T.; Toda, T.; Yanagida, M.; Hakoshima, T. Structural basis for the diversity of DNA recognition by bZip transcription factors. Nat. Struct. Biol. 2000, 7, 889–893. [Google Scholar] [PubMed]
- Garvie, C.W.; Wolberger, C. Recognition of specific DNA sequences. Mol. Cell. 2001, 8, 937–946. [Google Scholar] [CrossRef]
- Aramata, S.; Han, S.I.; Kataoka, K. Roles and regulation of transcription factor MafA in islet β-cells. Endocrinol. J. 2007, 54, 659–666. [Google Scholar] [CrossRef]
- Guo, S.; Burnette, R.; Zhao, L.; Vanderford, N.L.; Poitout, V.; Hagman, D.K.; Henderson, E.; Ozcan, S.; Wadzinski, B.E.; Stein, R. The stability and transactivation potential of the mammalian MafA transcription factor are regulated by serine 65 phosphorylation. J. Biol. Chem. 2009, 284, 759–765. [Google Scholar] [CrossRef] [PubMed]
- Benkhelifa, S.; Provot, S.; Nabais, E.; Eychene, A.; Calothy, G.; Felder-Schmittbuhl, M.P. Phosphorylation of MafA is essential for its transcriptional and biological properties. Mol. Cell. Biol. 2001, 21, 4441–4452. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, T.; Zhao, L.; Stein, R. The DNA binding activity of the RIPE3b1 transcription factor of insulin appears to be influenced by tyrosine phosphorylation. J. Biol. Chem. 2001, 276, 22071–22076. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Cissell, M.A.; Henderson, E.; Colbran, R.; Stein, R. The RIPE3b1 activator of the insulin gene is composed of a protein(s) of approximately 43 kDa, whose DNA binding activity is inhibited by protein phosphatase treatment. J. Biol. Chem. 2000, 275, 10532–10537. [Google Scholar] [CrossRef] [PubMed]
- Sii-Felice, K.; Pouponnot, C.; Gillet, S.; Lecoin, L.; Girault, J.A.; Eychene, A.; Felder-Schmittbuhl, M.P. MafA transcription factor is phosphorylated by p38 MAP kinase. FEBS Lett. 2005, 579, 3547–3554. [Google Scholar] [CrossRef] [PubMed]
- White, M.F. IRS proteins and the common path to diabetes. Am. J. Phys. Endocrinol. Metab. 2002, 283, E413–E422. [Google Scholar] [CrossRef] [PubMed]
- Lavan, B.E.; Fantin, V.R.; Chang, E.T.; Lane, W.S.; Keller, S.R.; Lienhard, G.E. A novel 160-kDa phosphotyrosine protein in insulin-treated embryonic kidney cells is a new member of the insulin receptor substrate family. J. Biol. Chem. 1997, 272, 21403–21407. [Google Scholar] [CrossRef] [PubMed]
- Dearth, R.K.; Cui, X.; Kim, H.J.; Hadsell, D.L.; Lee, A.V. Oncogenic transformation by the signaling adaptor proteins insulin receptor substrate (IRS)-1 and IRS-2. Cell Cycle 2007, 6, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, C.M.; Emanuelli, B.; Kahn, C.R. Critical nodes in signalling pathways: Insights into insulin action. Nat. Rev. Mol. Cell Biol. 2006, 7, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Hakuno, F.; Fukushima, T.; Yoneyama, Y.; Kamei, H.; Ozoe, A.; Yoshihara, H.; Yamanaka, D.; Shibano, T.; Sone-Yonezawa, M.; Yu, B.C.; et al. The novel functions of high-molecular-mass complexes containing insulin receptor substrates in mediation and modulation of insulin-like activities: Emerging concept of diverse functions by IRS-associated proteins. Front. Endocrinol. (Lausanne) 2015, 6, 73. [Google Scholar] [CrossRef] [PubMed]
- Ozoe, A.; Sone, M.; Fukushima, T.; Kataoka, N.; Arai, T.; Chida, K.; Asano, T.; Hakuno, F.; Takahashi, S. Insulin receptor substrate-1 (IRS-1) forms a ribonucleoprotein complex associated with polysomes. FEBS Lett. 2013, 587, 2319–2324. [Google Scholar] [CrossRef] [PubMed]
- Ozoe, A.; Sone, M.; Fukushima, T.; Kataoka, N.; Chida, K.; Asano, T.; Hakuno, F.; Takahashi, S. Insulin receptor substrate-1 associates with small nucleolar rna which contributes to ribosome biogenesis. Front. Endocrinol. (Lausanne) 2014, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Mardilovich, K.; Pankratz, S.L.; Shaw, L.M. Expression and function of the insulin receptor substrate proteins in cancer. Cell Commun. Signal. 2009, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Saltiel, A.R.; Kahn, C.R. Insulin signalling and the regulation of glucose and lipid metabolism. Nature 2001, 414, 799–806. [Google Scholar] [CrossRef] [PubMed]
- Rocchi, S.; Tartare-Deckert, S.; Sawka-Verhelle, D.; Gamha, A.; van Obberghen, E. Interaction of SH2-containing protein tyrosine phosphatase 2 with the insulin receptor and the insulin-like growth factor-I receptor: Studies of the domains involved using the yeast two-hybrid system. Endocrinology 1996, 137, 4944–4952. [Google Scholar] [CrossRef] [PubMed]
- Myers, M.G.; Mendez, R.; Shi, P.; Pierce, J.H.; Rhoads, R.; White, M.F. The cooh-terminal tyrosine phosphorylation sites on IRS-1 bind SHP-2 and negatively regulate insulin signaling. J. Biol. Chem. 1998, 273, 26908–26914. [Google Scholar] [CrossRef] [PubMed]
- Hanke, S.; Mann, M. The phosphotyrosine interactome of the insulin receptor family and its substrates IRS-1 and IRS-2. Mol. Cell. Proteom. 2009, 8, 519–534. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, K.; Delibegovic, M.; Matsuo, I.; Nagata, N.; Liu, S.M.; Bettaieb, A.; Xi, Y.N.; Araki, K.; Yang, W.T.; Kahn, B.B.; et al. Altered glucose homeostasis in mice with liver-specific deletion of Src homology phosphatase 2. J. Biol. Chem. 2010, 285, 39750–39758. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, S.; Wandless, T.J.; Shoelson, S.E.; Neel, B.G.; Walsh, C.T. Activation of the SH2-containing protein-tyrosine-phosphatase, SH-PTP2, by phosphotyrosine-containing peptides derived from insulin-receptor substrate-1. J. Biol. Chem. 1994, 269, 13614–13622. [Google Scholar] [PubMed]
- Pluskey, S.; Wandless, T.J.; Walsh, C.T.; Shoelson, S.E. Potent stimulation of SH-PTP2 phosphatase-activity by simultaneous occupancy of both SH2 domains. J. Biol. Chem. 1995, 270, 2897–2900. [Google Scholar] [CrossRef] [PubMed]
- Esposito, D.L.; Li, Y.; Cama, A.; Quon, M.J. Tyr(612) and tyr(632) in human insulin receptor substrate-1 are important for full activation of insulin-stimulated phosphatidylinositol 3-kinase activity and translocation of GLUT4 in adipose cells. Endocrinology 2001, 142, 2833–2840. [Google Scholar] [CrossRef] [PubMed]
- Cantley, L.C. The phosphoinositide 3-kinase pathway. Science 2002, 296, 1655–1657. [Google Scholar] [CrossRef] [PubMed]
- Boura-Halfon, S.; Zick, Y. Phosphorylation of IRS proteins, insulin action, and insulin resistance. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E581–E591. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gao, Z.; Yin, J.; Quon, M.J.; Ye, J. S6K directly phosphorylates IRS 1 on Ser-270 to promote insulin resistance in response to TNF-(α) signaling through IKK2. J. Biol. Chem. 2008, 283, 35375–35382. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Soos, T.J.; Li, X.; Wu, J.; Degennaro, M.; Sun, X.; Littman, D.R.; Birnbaum, M.J.; Polakiewicz, R.D. Protein kinase C theta inhibits insulin signaling by phosphorylating IRS1 at Ser(1101). J. Biol. Chem. 2004, 279, 45304–45307. [Google Scholar] [CrossRef] [PubMed]
- Petersen, J.; Wurzbacher, S.J.; Williamson, N.A.; Ramarathinam, S.H.; Reid, H.H.; Nair, A.K.N.; Zhao, A.Y.; Nastovska, R.; Rudge, G.; Rossjohn, J.; et al. Phosphorylated self-peptides alter human leukocyte antigen class I-restricted antigen presentation and generate tumor-specific epitopes. Proc.Natl. Acad. Sci. USA 2009, 106, 2776–2781. [Google Scholar] [CrossRef] [PubMed]
- Ikink, G.J.; Boer, M.; Bakker, E.R.; Hilkens, J. IRS4 induces mammary tumorigenesis and confers resistance to HER2-targeted therapy through constitutive PI3K/AKT-pathway hyperactivation. Nat. Commun. 2016, 7, 13567. [Google Scholar] [CrossRef] [PubMed]
- Herbst, J.J.; Andrews, G.; Contillo, L.; Lamphere, L.; Gardner, J.; Lienhard, G.E.; Gibbs, E.M. Potent activation of phosphatidylinositol 3’-kinase by simple phosphotyrosine peptides derived from insulin receptor substrate 1 containing two ymxm motifs for binding SH2 domains. Biochemistry 1994, 33, 9376–9381. [Google Scholar] [CrossRef] [PubMed]
- Brissova, M.; Blaha, M.; Spear, C.; Nicholson, W.; Radhika, A.; Shiota, M.; Charron, M.J.; Wright, C.V.; Powers, A.C. Reduced PDX-1 expression impairs islet response to insulin resistance and worsens glucose homeostasis. Am. J. Phys. Endocrinol. Metab. 2005, 288, E707–E714. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, R.N.; Jhala, U.S.; Winnay, J.N.; Krajewski, S.; Montminy, M.; Kahn, C.R. PDX-1 haploinsufficiency limits the compensatory islet hyperplasia that occurs in response to insulin resistance. J. Clin. Investig. 2004, 114, 828–836. [Google Scholar] [CrossRef] [PubMed]
- Holland, A.M.; Gonez, L.J.; Naselli, G.; Macdonald, R.J.; Harrison, L.C. Conditional expression demonstrates the role of the homeodomain transcription factor PDX1 in maintenance and regeneration of β-cells in the adult pancreas. Diabetes 2005, 54, 2586–2595. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, J.; Carlsson, L.; Edlund, T.; Edlund, H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 1994, 371, 606–609. [Google Scholar] [CrossRef] [PubMed]
- Hui, H.; Perfetti, R. Pancreas duodenum homeobox-1 regulates pancreas development during embryogenesis and islet cell function in adulthood. Eur. J. Endocrinol. 2002, 146, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Miller, C.; Habener, J.F. Functional regions of the homeodomain protein IDX-1 required for transactivation of the rat somatostatin gene. Endocrinology 1996, 137, 2959–2967. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Desai, B.M.; Stoffers, D.A. Identification of PCIF1, a POZ domain protein that inhibits PDX-1 (MODY4) transcriptional activity. Mol. Cell. Biol. 2004, 24, 4372–4383. [Google Scholar] [CrossRef] [PubMed]
- An, R.; da Silva Xavier, G.; Semplici, F.; Vakhshouri, S.; Hao, H.X.; Rutter, J.; Pagano, M.A.; Meggio, F.; Pinna, L.A.; Rutter, G.A. Pancreatic and duodenal homeobox 1 (PDX1) phosphorylation at Serine-269 is HIPK2-dependent and affects PDX1 subnuclear localization. Biochem. Biophys. Res. Commun. 2010, 399, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Gehring, W.J.; Affolter, M.; Burglin, T. Homeodomain proteins. Annu. Rev. Biochem. 1994, 63, 487–526. [Google Scholar] [CrossRef] [PubMed]
- Hoey, T.; Levine, M. Divergent homeo box proteins recognize similar DNA sequences in drosophila. Nature 1988, 332, 858–861. [Google Scholar] [CrossRef] [PubMed]
- Smock, R.G.; Gierasch, L.M. Sending signals dynamically. Science 2009, 324, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Dunker, A.K. Understanding protein non-folding. Biochim. Biophys. Acta 2010, 1804, 1231–1264. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Vanella, L.; Inoue, K.; Burgess, A.; Gotlinger, K.; Manthati, V.L.; Koduru, S.R.; Zeldin, D.C.; Falck, J.R.; Schwartzman, M.L.; et al. Epoxyeicosatrienoic acid agonist regulates human mesenchymal stem cell-derived adipocytes through activation of HO-1-PAKT signaling and a decrease in PPARγ. Stem. Cells Dev. 2010, 19, 1863–1873. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Fu, Z.; Liu, Z. Adiponectin and insulin cross talk: The microvascular connection. Trends Cardiovasc. Med. 2014, 24, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Shibata, R.; Sato, K.; Pimentel, D.R.; Takemura, Y.; Kihara, S.; Ohashi, K.; Funahashi, T.; Ouchi, N.; Walsh, K. Adiponectin protects against myocardial ischemia-reperfusion injury through AMPK- and COX-2-dependent mechanisms. Nat. Med. 2005, 11, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Li, F.Y.; Cheng, K.K.; Lam, K.S.; Vanhoutte, P.M.; Xu, A. Cross-talk between adipose tissue and vasculature: Role of adiponectin. Acta Physiol. 2011, 203, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Misu, H.; Ishikura, K.; Kurita, S.; Takeshita, Y.; Ota, T.; Saito, Y.; Takahashi, K.; Kaneko, S.; Takamura, T. Inverse correlation between serum levels of selenoprotein P and adiponectin in patients with type 2 diabetes. PLoS ONE 2012, 7, e34952. [Google Scholar] [CrossRef] [PubMed]
- Takeda, Y.; Nakanishi, K.; Tachibana, I.; Kumanogoh, A. Adiponectin: A novel link between adipocytes and copd. Vitam. Horm. 2012, 90, 419–435. [Google Scholar] [PubMed]
- Lau, W.B.; Tao, L.; Wang, Y.; Li, R.; Ma, X.L. Systemic adiponectin malfunction as a risk factor for cardiovascular disease. Antioxid. Redox Signal. 2011, 15, 1863–1873. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, B.J.; Scalia, R.G.; Ma, X.L. Protective vascular and myocardial effects of adiponectin. Nat. Clin. Pract. Cardiovasc. Med. 2009, 6, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Min, X.; Lemon, B.; Tang, J.; Liu, Q.; Zhang, R.; Walker, N.; Li, Y.; Wang, Z. Crystal structure of a single-chain trimer of human adiponectin globular domain. FEBS Lett. 2012, 586, 912–917. [Google Scholar] [CrossRef] [PubMed]
- Waki, H.; Yamauchi, T.; Kamon, J.; Ito, Y.; Uchida, S.; Kita, S.; Hara, K.; Hada, Y.; Vasseur, F.; Froguel, P.; et al. Impaired multimerization of human adiponectin mutants associated with diabetes. Molecular structure and multimer formation of adiponectin. J. Biol. Chem. 2003, 278, 40352–40363. [Google Scholar] [CrossRef] [PubMed]
- Kuchay, S.; Duan, S.; Schenkein, E.; Peschiaroli, A.; Saraf, A.; Florens, L.; Washburn, M.P.; Pagano, M. FBXL2- and PTPL1-mediated degradation of p110-free p85β regulatory subunit controls the PI(3)K signalling cascade. Nat. Cell Biol. 2013, 15, 472–480. [Google Scholar] [CrossRef] [PubMed]
- Chalhoub, N.; Baker, S.J. Pten and the PI3-kinase pathway in cancer. Annu. Rev. Pathol. 2009, 4, 127–150. [Google Scholar] [CrossRef] [PubMed]
- Thorpe, L.M.; Yuzugullu, H.; Zhao, J.J. PI3K in cancer: Divergent roles of isoforms, modes of activation and therapeutic targeting. Nat. Rev. Cancer 2015, 15, 7–24. [Google Scholar] [CrossRef] [PubMed]
- Vanhaesebroeck, B.; Guillermet-Guibert, J.; Graupera, M.; Bilanges, B. The emerging mechanisms of isoform-specific PI3K signalling. Nat. Rev. Mol. Cell. Biol. 2010, 11, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Fruman, D.A.; Cantley, L.C.; Carpenter, C.L. Structural organization and alternative splicing of the murine phosphoinositide 3-kinase p85 α gene. Genomics 1996, 37, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Ueki, K.; Fruman, D.A.; Brachmann, S.M.; Tseng, Y.H.; Cantley, L.C.; Kahn, C.R. Molecular balance between the regulatory and catalytic subunits of phosphoinositide 3-kinase regulates cell signaling and survival. Mol. Cell. Biol. 2002, 22, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Ueki, K.; Yballe, C.M.; Brachmann, S.M.; Vicent, D.; Watt, J.M.; Kahn, C.R.; Cantley, L.C. Increased insulin sensitivity in mice lacking p85β subunit of phosphoinositide 3-kinase. Proc. Natl. Acad. Sci. USA 2002, 99, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Brachmann, S.M.; Ueki, K.; Engelman, J.A.; Kahn, R.C.; Cantley, L.C. Phosphoinositide 3-kinase catalytic subunit deletion and regulatory subunit deletion have opposite effects on insulin sensitivity in mice. Mol. Cell. Biol. 2005, 25, 1596–1607. [Google Scholar] [CrossRef] [PubMed]
- Mauvais-Jarvis, F.; Ueki, K.; Fruman, D.A.; Hirshman, M.F.; Sakamoto, K.; Goodyear, L.J.; Iannacone, M.; Accili, D.; Cantley, L.C.; Kahn, C.R. Reduced expression of the murine p85α subunit of phosphoinositide 3-kinase improves insulin signaling and ameliorates diabetes. J. Clin. Investig. 2002, 109, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Park, S.W.; Zhou, Y.; Lee, J.; Lu, A.; Sun, C.; Chung, J.; Ueki, K.; Ozcan, U. The regulatory subunits of PI3K, p85 α and p85β, interact with xbp-1 and increase its nuclear translocation. Nat. Med. 2010, 16, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Virkamaki, A.; Ueki, K.; Kahn, C.R. Protein-protein interaction in insulin signaling and the molecular mechanisms of insulin resistance. J. Clin. Investig. 1999, 103, 931–943. [Google Scholar] [CrossRef] [PubMed]
- Straub, S.G.; Sharp, G.W. Glucose-dependent insulinotropic polypeptide stimulates insulin secretion via increased cyclic AMP and [Ca2+]i and a wortmannin-sensitive signalling pathway. Biochem. Biophys. Res. Commun. 1996, 224, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Straub, S.G.; Sharp, G.W. A wortmannin-sensitive signal transduction pathway is involved in the stimulation of insulin release by vasoactive intestinal polypeptide and pituitary adenylate cyclase-activating polypeptide. J. Biol. Chem. 1996, 271, 1660–1668. [Google Scholar] [CrossRef] [PubMed]
- Hagiwara, S.; Sakurai, T.; Tashiro, F.; Hashimoto, Y.; Matsuda, Y.; Nonomura, Y.; Miyazaki, J. An inhibitory role for phosphatidylinositol 3-kinase in insulin secretion from pancreatic B cell line MIN6. Biochem. Biophys. Res. Commun. 1995, 214, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Eto, K.; Yamashita, T.; Tsubamoto, Y.; Terauchi, Y.; Hirose, K.; Kubota, N.; Yamashita, S.; Taka, J.; Satoh, S.; Sekihara, H.; et al. Phosphatidylinositol 3-kinase suppresses glucose-stimulated insulin secretion by affecting post-cytosolic [Ca2+] elevation signals. Diabetes 2002, 51, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Chen, M.; Yan, B.; He, X.; Chen, X.; Li, D. Identification of a role for the PI3K/AKT/MTOR signaling pathway in innate immune cells. PLoS ONE 2014, 9, e94496. [Google Scholar] [CrossRef] [PubMed]
- Koyasu, S. The role of PI3K in immune cells. Nat. Immunol. 2003, 4, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Ruckle, T.; Schwarz, M.K.; Rommel, C. PI3Kγ inhibition: Towards an ‘aspirin of the 21st century’? Nat. Rev. Drug Discov. 2006, 5, 903–918. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, P.E.; Joseph, J.W.; Yau, D.; Diao, J.; Asghar, Z.; Dai, F.; Oudit, G.Y.; Patel, M.M.; Backx, P.H.; Wheeler, M.B. Impaired glucose-stimulated insulin secretion, enhanced intraperitoneal insulin tolerance, and increased β-cell mass in mice lacking the p110γ isoform of phosphoinositide 3-kinase. Endocrinology 2004, 145, 4078–4083. [Google Scholar] [CrossRef] [PubMed]
- Li, L.X.; MacDonald, P.E.; Ahn, D.S.; Oudit, G.Y.; Backx, P.H.; Brubaker, P.L. Role of phosphatidylinositol 3-kinasegamma in the β-cell: Interactions with glucagon-like peptide-1. Endocrinology 2006, 147, 3318–3325. [Google Scholar] [CrossRef] [PubMed]
- Pigeau, G.M.; Kolic, J.; Ball, B.J.; Hoppa, M.B.; Wang, Y.W.; Ruckle, T.; Woo, M.; Manning Fox, J.E.; MacDonald, P.E. Insulin granule recruitment and exocytosis is dependent on p110γ in insulinoma and human β-cells. Diabetes 2009, 58, 2084–2092. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Chen, J.K.; Schreiber, S.T.; Clardy, J. Crystal structure of P13K SH3 domain at 20 angstroms resolution. J. Mol. Biol. 1996, 257, 632–643. [Google Scholar] [CrossRef] [PubMed]
- Hoedemaeker, F.J.; Siegal, G.; Roe, S.M.; Driscoll, P.C.; Abrahams, J.P. Crystal structure of the C-terminal SH2 domain of the p85α regulatory subunit of phosphoinositide 3-kinase: An SH2 domain mimicking its own substrate. J. Mol. Biol. 1999, 292, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Nolte, R.T.; Eck, M.J.; Schlessinger, J.; Shoelson, S.E.; Harrison, S.C. Crystal structure of the π 3-kinase p85 amino-terminal SH2 domain and its phosphopeptide complexes. Nat. Struct. Biol. 1996, 3, 364–374. [Google Scholar] [CrossRef] [PubMed]
- Inukai, K.; Funaki, M.; Anai, M.; Ogihara, T.; Katagiri, H.; Fukushima, Y.; Sakoda, H.; Onishi, Y.; Ono, H.; Fujishiro, M.; et al. Five isoforms of the phosphatidylinositol 3-kinase regulatory subunit exhibit different associations with receptor tyrosine kinases and their tyrosine phosphorylations. FEBS Lett. 2001, 490, 32–38. [Google Scholar] [CrossRef]
- Schauder, C.; Ma, L.C.; Krug, R.M.; Montelione, G.T.; Guan, R. Structure of the iSH2 domain of human phosphatidylinositol 3-kinase p85β subunit reveals conformational plasticity in the interhelical turn region. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 1567–1571. [Google Scholar] [CrossRef] [PubMed]
- Dhand, R.; Hiles, I.; Panayotou, G.; Roche, S.; Fry, M.J.; Gout, I.; Totty, N.F.; Truong, O.; Vicendo, P.; Yonezawa, K.; et al. π 3-kinase is a dual specificity enzyme: Autoregulation by an intrinsic protein-serine kinase activity. EMBO J. 1994, 13, 522–533. [Google Scholar] [PubMed]
- Fu, Z.; Aronoff-Spencer, E.; Wu, H.Y.; Gerfen, G.J.; Backer, J.M. The iSH2 domain of π 3-kinase is a rigid tether for p110 and not a conformational switch. Arch. Biochem. Biophys. 2004, 432, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Elis, W.; Lessmann, E.; Oelgeschlager, M.; Huber, M. Mutations in the inter-SH2 domain of the regulatory subunit of phosphoinositide 3-kinase: Effects on catalytic subunit binding and holoenzyme function. Biol. Chem. 2006, 387, 1567–1573. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.H.; Mandelker, D.; Schmidt-Kittler, O.; Samuels, Y.; Velculescu, V.E.; Kinzler, K.W.; Vogelstein, B.; Gabelli, S.B.; Amzel, L.M. The structure of a human p110α/p85α complex elucidates the effects of oncogenic PI3Kα mutations. Science 2007, 318, 1744–1748. [Google Scholar] [CrossRef] [PubMed]
- Gazi, A.D.; Bastaki, M.; Charova, S.N.; Gkougkoulia, E.A.; Kapellios, E.A.; Panopoulos, N.J.; Kokkinidis, M. Evidence for a coiled-coil interaction mode of disordered proteins from bacterial type Ⅲ secretion systems. J. Biol. Chem. 2008, 283, 34062–34068. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, R.M.; Sykes, B.D. Isoform-specific variation in the intrinsic disorder of troponin Ⅰ. Proteins 2008, 73, 338–350. [Google Scholar] [CrossRef] [PubMed]
- Szappanos, B.; Suveges, D.; Nyitray, L.; Perczel, A.; Gaspari, Z. Folded-unfolded cross-predictions and protein evolution: The case study of coiled-coils. FEBS Lett. 2010, 584, 1623–1627. [Google Scholar] [CrossRef] [PubMed]
- Peysselon, F.; Xue, B.; Uversky, V.N.; Ricard-Blum, S. Intrinsic disorder of the extracellular matrix. Mol. Biosyst. 2011, 7, 3353–3365. [Google Scholar] [CrossRef] [PubMed]
- Anurag, M.; Singh, G.P.; Dash, D. Location of disorder in coiled coil proteins is influenced by its biological role and subcellular localization: A go-based study on human proteome. Mol. Biosyst. 2012, 8, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Gaspari, Z. Is five percent too small? Analysis of the overlaps between disorder, coiled coil and collagen predictions in complete proteomes. Proteomes 2014, 2, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Tang, H.; Leng, J.; Jiang, Q. Suppressors of cytokine signaling (SOCS) and type 2 diabetes. Mol. Biol. Rep. 2014, 41, 2265–2274. [Google Scholar] [CrossRef] [PubMed]
- Hilton, D.J.; Richardson, R.T.; Alexander, W.S.; Viney, E.M.; Willson, T.A.; Sprigg, N.S.; Starr, R.; Nicholson, S.E.; Metcalf, D.; Nicola, N.A. Twenty proteins containing a C-terminal SOCS box form five structural classes. Proc. Natl. Acad. Sci. USA 1998, 95, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Waiboci, L.W.; Ahmed, C.M.; Mujtaba, M.G.; Flowers, L.O.; Martin, J.P.; Haider, M.I.; Johnson, H.M. Both the suppressor of cytokine signaling 1 (SOCS-1) kinase inhibitory region and SOCS-1 mimetic bind to JAK2 autophosphorylation site: Implications for the development of a SOCS-1 antagonist. J. Immunol. 2007, 178, 5058–5068. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, A.; Yasukawa, H.; Suzuki, A.; Kamizono, S.; Syoda, T.; Kinjyo, I.; Sasaki, M.; Johnston, J.A.; Yoshimura, A. Cytokine-inducible SH2 protein-3 (CIS3/SOCS3) inhibits janus tyrosine kinase by binding through the N-terminal kinase inhibitory region as well as SH2 domain. Genes Cells 1999, 4, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Yasukawa, H.; Misawa, H.; Sakamoto, H.; Masuhara, M.; Sasaki, A.; Wakioka, T.; Ohtsuka, S.; Imaizumi, T.; Matsuda, T.; Ihle, J.N.; et al. The JAK-binding protein JAB inhibits janus tyrosine kinase activity through binding in the activation loop. EMBO J. 1999, 18, 1309–1320. [Google Scholar] [CrossRef] [PubMed]
- Babon, J.J.; McManus, E.J.; Yao, S.; DeSouza, D.P.; Mielke, L.A.; Sprigg, N.S.; Willson, T.A.; Hilton, D.J.; Nicola, N.A.; Baca, M.; et al. The structure of SOCS3 reveals the basis of the extended SH2 domain function and identifies an unstructured insertion that regulates stability. Mol. Cell. 2006, 22, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Bergamin, E.; Wu, J.; Hubbard, S.R. Structural basis for phosphotyrosine recognition by suppressor of cytokine signaling-3. Structure 2006, 14, 1285–1292. [Google Scholar] [CrossRef] [PubMed]
- Kershaw, N.J.; Murphy, J.M.; Liau, N.P.D.; Varghese, L.N.; Laktyushin, A.; Whitlock, E.L.; Lucet, I.S.; Nicola, N.A.; Babon, J.J. SOCS3 binds specific receptor-JAK complexes to control cytokine signaling by direct kinase inhibition. Nat. Struct. Mol. Biol. 2013, 20, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Babon, J.J.; Yao, S.; DeSouza, D.P.; Harrison, C.F.; Fabri, L.J.; Liepinsh, E.; Scrofani, S.D.; Baca, M.; Norton, R.S. Secondary structure assignment of mouse SOCS3 by NMR defines the domain boundaries and identifies an unstructured insertion in the SH2 domain. FEBS J. 2005, 272, 6120–6130. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.J.; Munro, K.M.; Palmer, T.M. Role of ubiquitylation in controlling suppressor of cytokine signalling 3 (SOCS3) function and expression. Cells 2014, 3, 546–562. [Google Scholar] [CrossRef] [PubMed]
- Jager, L.D.; Dabelic, R.; Waiboci, L.W.; Lau, K.; Haider, M.S.; Ahmed, C.M.I.; Larkin, J.; David, S.; Johnson, H.M. The kinase inhibitory region of SOCS-1 is sufficient to inhibit T-helper 17 and other immune functions in experimental allergic encephalomyelitis. J. Neuroimmunol. 2011, 232, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, C.M.; Larkin, J., 3rd; Johnson, H.M. SOCS1 mimetics and antagonists: A complementary approach to positive and negative regulation of immune function. Front. Immunol. 2015, 6, 183. [Google Scholar] [CrossRef] [PubMed]
- Babon, J.J.; Sabo, J.K.; Soetopo, A.; Yao, S.; Bailey, M.F.; Zhang, J.G.; Nicola, N.A.; Norton, R.S. The socs box domain of SOCS3: Structure and interaction with the elonginbc-cullin5 ubiquitin ligase. J. Mol. Biol. 2008, 381, 928–940. [Google Scholar] [CrossRef] [PubMed]
- Redwan, R.M.; Matar, S.M.; El-Aziz, G.A.; Serour, E.A. Synthesis of the human insulin gene: Protein expression, scaling up and bioactivity. Prep. Biochem. Biotechnol. 2008, 38, 24–39. [Google Scholar] [CrossRef] [PubMed]
- Wild, S.; Roglic, G.; Green, A.; Sicree, R.; King, H. Global prevalence of diabetes: Estimates for the year 2000 and projections for 2030. Diabetes Care 2004, 27, 1047–1053. [Google Scholar] [CrossRef] [PubMed]
- Sonksen, P.; Sonksen, J. Insulin: Understanding its action in health and disease. Br. J. Anaesth. 2000, 85, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Dimitriadis, G.; Mitrou, P.; Lambadiari, V.; Maratou, E.; Raptis, S.A. Insulin effects in muscle and adipose tissue. Diabetes Res. Clin. Pract. 2011, 93 (Suppl. 1), S52–S59. [Google Scholar] [CrossRef]
- Koeslag, J.H.; Saunders, P.T.; Terblanche, E. A reappraisal of the blood glucose homeostat which comprehensively explains the type 2 diabetes mellitus-syndrome x complex. J. Physiol. 2003, 549, 333–346. [Google Scholar] [CrossRef] [PubMed]
- Szablewski, L. Diabetes mellitus: Influences on cancer risk. Diabetes Metab. Res. Rev. 2014, 30, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Suh, S.; Kim, K.W. Diabetes and cancer: Is diabetes causally related to cancer? Diabetes Metab. J. 2011, 35, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Vigneri, P.; Frasca, F.; Sciacca, L.; Pandini, G.; Vigneri, R. Diabetes and cancer. Endocr. Relat. Cancer 2009, 16, 1103–1123. [Google Scholar] [CrossRef] [PubMed]
- Pisani, P. Hyper-insulinaemia and cancer, meta-analyses of epidemiological studies. Arch. Physiol. Biochem. 2008, 114, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Frasca, F.; Pandini, G.; Scalia, P.; Sciacca, L.; Mineo, R.; Costantino, A.; Goldfine, I.D.; Belfiore, A.; Vigneri, R. Insulin receptor isoform A, a newly recognized, high-affinity insulin-like growth factor Ⅱ receptor in fetal and cancer cells. Mol. Cell. Biol. 1999, 19, 3278–3288. [Google Scholar] [CrossRef] [PubMed]
- Pollak, M. Insulin and insulin-like growth factor signalling in neoplasia. Nat. Rev. Cancer 2008, 8, 915–928. [Google Scholar] [CrossRef] [PubMed]
- Papa, V.; Pezzino, V.; Costantino, A.; Belfiore, A.; Giuffrida, D.; Frittitta, L.; Vannelli, G.B.; Brand, R.; Goldfine, I.D.; Vigneri, R. Elevated insulin receptor content in human breast cancer. J. Clin. Investig. 1990, 86, 1503–1510. [Google Scholar] [CrossRef] [PubMed]
- Giovannucci, E. Insulin, insulin-like growth factors and colon cancer: A review of the evidence. J. Nutr. 2001, 131, 3109S–3120S. [Google Scholar] [PubMed]
- Volkers, N. Diabetes and cancer: Scientists search for a possible link. J. Natl. Cancer Inst. 2000, 92, 192–194. [Google Scholar] [CrossRef] [PubMed]
- Redwan, E.M.; Linjawi, M.H.; Uversky, V.N. Looking at the carcinogenicity of human insulin analogues via the intrinsic disorder prism. Sci. Rep. 2016, 6, 23320. [Google Scholar] [CrossRef] [PubMed]
- Shabanpoor, F.; Separovic, F.; Wade, J.D. The human insulin superfamily of polypeptide hormones. Vitam. Horm. 2009, 80, 1–31. [Google Scholar] [PubMed]
- Emdin, S.O.; Dodson, G.G.; Cutfield, J.M.; Cutfield, S.M. Role of zinc in insulin biosynthesis. Some possible zinc-insulin interactions in the pancreatic B-cell. Diabetologia 1980, 19, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Bryant, C.; Spencer, D.B.; Miller, A.; Bakaysa, D.L.; McCune, K.S.; Maple, S.R.; Pekar, A.H.; Brems, D.N. Acid stabilization of insulin. Biochemistry 1993, 32, 8075–8082. [Google Scholar] [CrossRef] [PubMed]
- Waugh, D.F. A fibrous modification of insulin. I. The heat precipitate of insulin. J. Am. Chem. Soc. 1946, 68, 247–250. [Google Scholar] [CrossRef]
- Waugh, D.F.; Wilhelmson, D.F.; Commerford, S.L.; Sackler, M.L. Studies of the nucleation and growth reactions of selected types of insulin fibrils. J. Am. Chem. Soc. 1953, 75, 2592–2600. [Google Scholar] [CrossRef]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001, 19, 26–59. [Google Scholar] [CrossRef]
- Rose, A.S.; Hildebrand, P.W. Ngl viewer: A web application for molecular visualization. Nucleic Acids Res. 2015, 43, W576–W579. [Google Scholar] [CrossRef] [PubMed]
- De Meyts, P.; Whittaker, J. Structural biology of insulin and IGF1 receptors: Implications for drug design. Nat. Rev. Drug. Discov. 2002, 1, 769–783. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, S.R. The insulin receptor: Both a prototypical and atypical receptor tyrosine kinase. Cold Spring Harb. Perspect. Biol. 2013, 5, a008946. [Google Scholar] [CrossRef] [PubMed]
- Croll, T.I.; Smith, B.J.; Margetts, M.B.; Whittaker, J.; Weiss, M.A.; Ward, C.W.; Lawrence, M.C. Higher-resolution structure of the human insulin receptor ectodomain: Multi-modal inclusion of the insert domain. Structure 2016, 24, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Bedinger, D.H.; Adams, S.H. Metabolic, anabolic, and mitogenic insulin responses: A tissue-specific perspective for insulin receptor activators. Mol. Cell. Endocrinol. 2015, 415, 143–156. [Google Scholar] [CrossRef] [PubMed]
- De Meyts, P. Insulin/receptor binding: The last piece of the puzzle? What recent progress on the structure of the insulin/receptor complex tells us (or not) about negative cooperativity and activation. Bioessays 2015, 37, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Tatulian, S.A. Structural dynamics of insulin receptor and transmembrane signaling. Biochemistry 2015, 54, 5523–5532. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, R.; Haeri, A.; Dargahi, L.; Mohamed, Z.; Ahmadiani, A. Insulin in the brain: Sources, localization and functions. Mol. Neurobiol. 2013, 47, 145–171. [Google Scholar] [CrossRef] [PubMed]
- Kleinridders, A.; Ferris, H.A.; Cai, W.; Kahn, C.R. Insulin action in brain regulates systemic metabolism and brain function. Diabetes 2014, 63, 2232–2243. [Google Scholar] [CrossRef] [PubMed]
- Gralle, M. The neuronal insulin receptor in its environment. J. Neurochem. 2017, 140, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Adamo, M.; Raizada, M.K.; LeRoith, D. Insulin and insulin-like growth factor receptors in the nervous system. Mol. Neurobiol. 1989, 3, 71–100. [Google Scholar] [CrossRef] [PubMed]
- LeRoith, D.; Werner, H.; Faria, T.N.; Kato, H.; Adamo, M.; Roberts, C.T., Jr. Insulin-like growth factor receptors. Implications for nervous system function. Ann. N. Y. Acad. Sci. 1993, 692, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Moller, D.E.; Yokota, A.; Caro, J.F.; Flier, J.S. Tissue-specific expression of two alternatively spliced insulin receptor mRNAs in man. Mol. Endocrinol. 1989, 3, 1263–1269. [Google Scholar] [CrossRef] [PubMed]
- Craft, S. Alzheimer disease: Insulin resistance and AD–extending the translational path. Nat. Rev. Neurol. 2012, 8, 360–362. [Google Scholar] [CrossRef] [PubMed]
- Frasca, F.; Pandini, G.; Sciacca, L.; Pezzino, V.; Squatrito, S.; Belfiore, A.; Vigneri, R. The role of insulin receptors and IGF-I receptors in cancer and other diseases. Arch. Physiol. Biochem. 2008, 114, 23–37. [Google Scholar] [CrossRef] [PubMed]
- McKern, N.M.; Lawrence, M.C.; Streltsov, V.A.; Lou, M.Z.; Adams, T.E.; Lovrecz, G.O.; Elleman, T.C.; Richards, K.M.; Bentley, J.D.; Pilling, P.A.; et al. Structure of the insulin receptor ectodomain reveals a folded-over conformation. Nature 2006, 443, 218–221. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Maji, S.; Sanghera, N.; Gopalasingam, P.; Gorbunov, E.; Tarasov, S.; Epstein, O.; Klein-Seetharaman, J. Structure and dynamics of the insulin receptor: Implications for receptor activation and drug discovery. Drug Discov. Today 2017, 22, 1092–1102. [Google Scholar] [CrossRef] [PubMed]
- Lukman, S.; Al Safar, H.; Mun Lee, S.; Sim, K. Harnessing structural data of insulin and insulin receptor for therapeutic designs. J. Endocrinol. Metab. 2015, 5, 273–283. [Google Scholar] [CrossRef]
- Lou, M.; Garrett, T.P.; McKern, N.M.; Hoyne, P.A.; Epa, V.C.; Bentley, J.D.; Lovrecz, G.O.; Cosgrove, L.J.; Frenkel, M.J.; Ward, C.W. The first three domains of the insulin receptor differ structurally from the insulin-like growth factor 1 receptor in the regions governing ligand specificity. Proc. Natl. Acad. Sci. USA 2006, 103, 12429–12434. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wong, Y.L.; Kang, C. Solution structure of the transmembrane domain of the insulin receptor in detergent micelles. Biochim. Biophys. Acta 2014, 1838, 1313–1321. [Google Scholar] [CrossRef] [PubMed]
- Cabail, M.Z.; Li, S.; Lemmon, E.; Bowen, M.E.; Hubbard, S.R.; Miller, W.T. The insulin and IGF1 receptor kinase domains are functional dimers in the activated state. Nat. Commun. 2015, 6, 6406. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.J.; Huang, K.; Kong, G.; Chan, S.J.; Nakagawa, S.; Menting, J.G.; Hu, S.Q.; Whittaker, J.; Steiner, D.F.; Katsoyannis, P.G.; et al. Structural resolution of a tandem hormone-binding element in the insulin receptor and its implications for design of peptide agonists. Proc. Natl. Acad. Sci. USA 2010, 107, 6771–6776. [Google Scholar] [CrossRef] [PubMed]
- Prilusky, J.; Felder, C.E.; Zeev-Ben-Mordehai, T.; Rydberg, E.H.; Man, O.; Beckmann, J.S.; Silman, I.; Sussman, J.L. Foldindex: A simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics 2005, 21, 3435–3438. [Google Scholar] [CrossRef] [PubMed]
- Campen, A.; Williams, R.M.; Brown, C.J.; Meng, J.; Uversky, V.N.; Dunker, A.K. TOP-IDP-scale: A new amino acid scale measuring propensity for intrinsic disorder. Protein Pept. Lett. 2008, 15, 956–963. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Kurgan, L. Accurate prediction of disorder in protein chains with a comprehensive and empirically designed consensus. J. Biomol. Struct. Dyn. 2014, 32, 448–464. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.L.; Kurgan, L. Comprehensive comparative assessment of in-silico predictors of disordered regions. Curr. Protein Pept. Sci. 2012, 13, 6–18. [Google Scholar] [CrossRef] [PubMed]
- Walsh, I.; Giollo, M.; Di Domenico, T.; Ferrari, C.; Zimmermann, O.; Tosatto, S.C. Comprehensive large-scale assessment of intrinsic protein disorder. Bioinformatics 2015, 31, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Kurgan, L. On the complementarity of the consensus-based disorder prediction. Pac. Symp. Biocomput. 2012, 176–187. [Google Scholar]
- Xue, B.; Oldfield, C.J.; Dunker, A.K.; Uversky, V.N. Cdf it all: Consensus prediction of intrinsically disordered proteins based on various cumulative distribution functions. FEBS Lett. 2009, 583, 1469–1474. [Google Scholar] [CrossRef] [PubMed]
- Mohan, A.; Sullivan, W.J., Jr.; Radivojac, P.; Dunker, A.K.; Uversky, V.N. Intrinsic disorder in pathogenic and non-pathogenic microbes: Discovering and analyzing the unfoldomes of early-branching eukaryotes. Mol. Biosyst. 2008, 4, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Oldfield, C.J.; Meng, J.; Hsu, W.-L.; Xue, B.; Uversky, V.N.; Romero, P.; Dunker, A.K. Subclassifying disordered proteins by the CH-CDF plot method. Pac. Symp. Biocomput. 2012, 128–139. [Google Scholar]
- Malhis, N.; Jacobson, M.; Gsponer, J. MoRF chibi SYSTEM: Software tools for the identification of MoRFs in protein sequences. Nucleic Acids Res. 2016, 44, W488–W493. [Google Scholar] [CrossRef] [PubMed]



















© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, Z.; Uversky, V.N. A Comprehensive Survey of the Roles of Highly Disordered Proteins in Type 2 Diabetes. Int. J. Mol. Sci. 2017, 18, 2010. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18102010
Du Z, Uversky VN. A Comprehensive Survey of the Roles of Highly Disordered Proteins in Type 2 Diabetes. International Journal of Molecular Sciences. 2017; 18(10):2010. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18102010
Chicago/Turabian StyleDu, Zhihua, and Vladimir N. Uversky. 2017. "A Comprehensive Survey of the Roles of Highly Disordered Proteins in Type 2 Diabetes" International Journal of Molecular Sciences 18, no. 10: 2010. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18102010