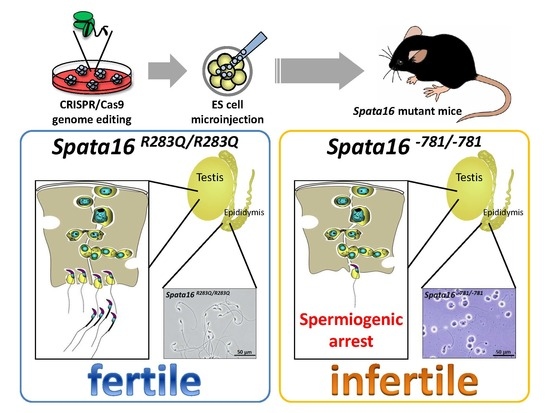
Human Globozoospermia-Related Gene Spata16 Is Required for Sperm Formation Revealed by CRISPR/Cas9-Mediated Mouse Models
Abstract
:1. Introduction
2. Results
2.1. Testis-Specific Expression and Protein Sequence Alignment of Mouse SPATA16
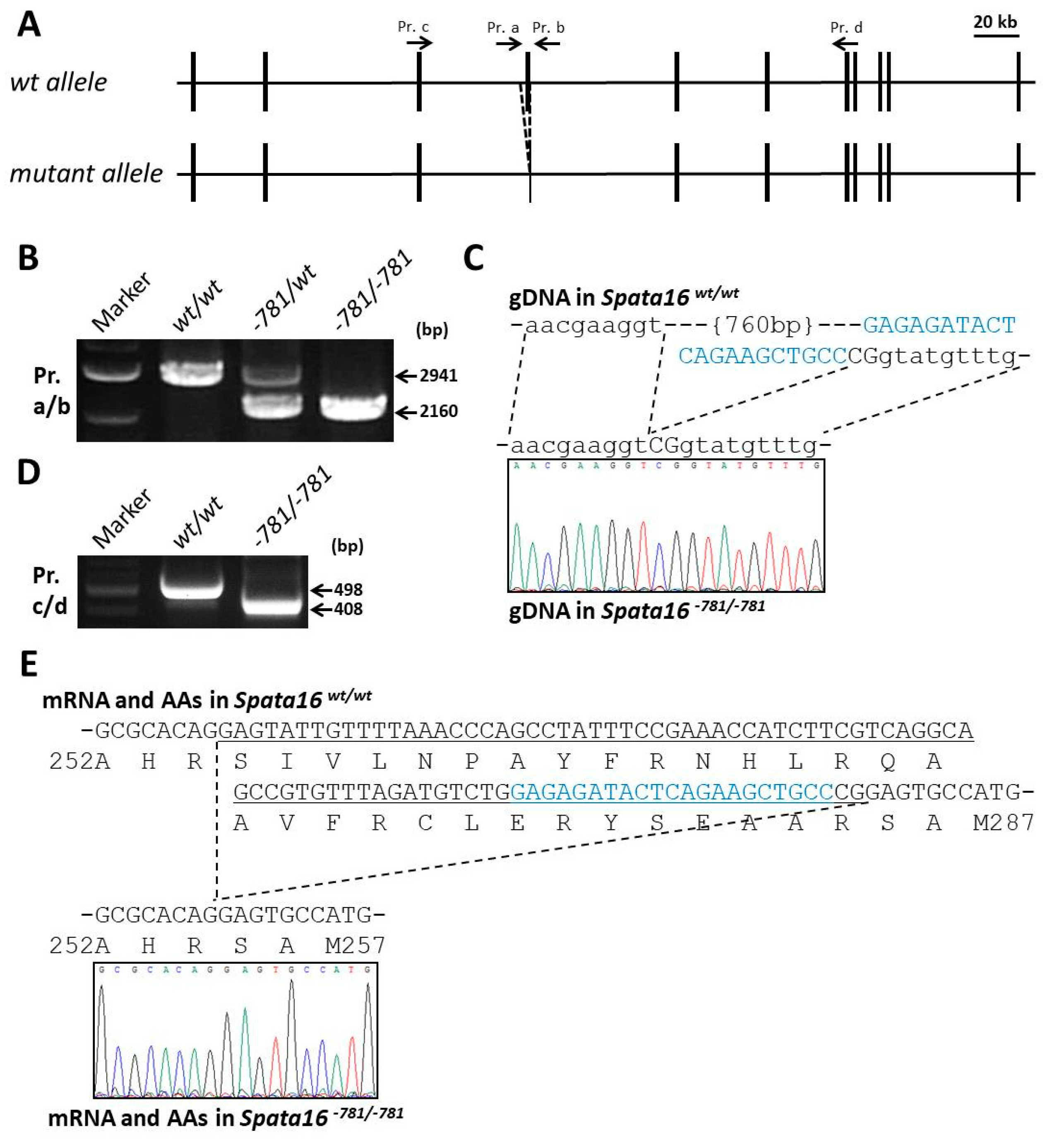
2.2. Targeted Point Mutation and Deletion of Spata16 by CRISPR/Cas9
2.3. Fertilizing Ability of Spata16 Mutant Male Mice
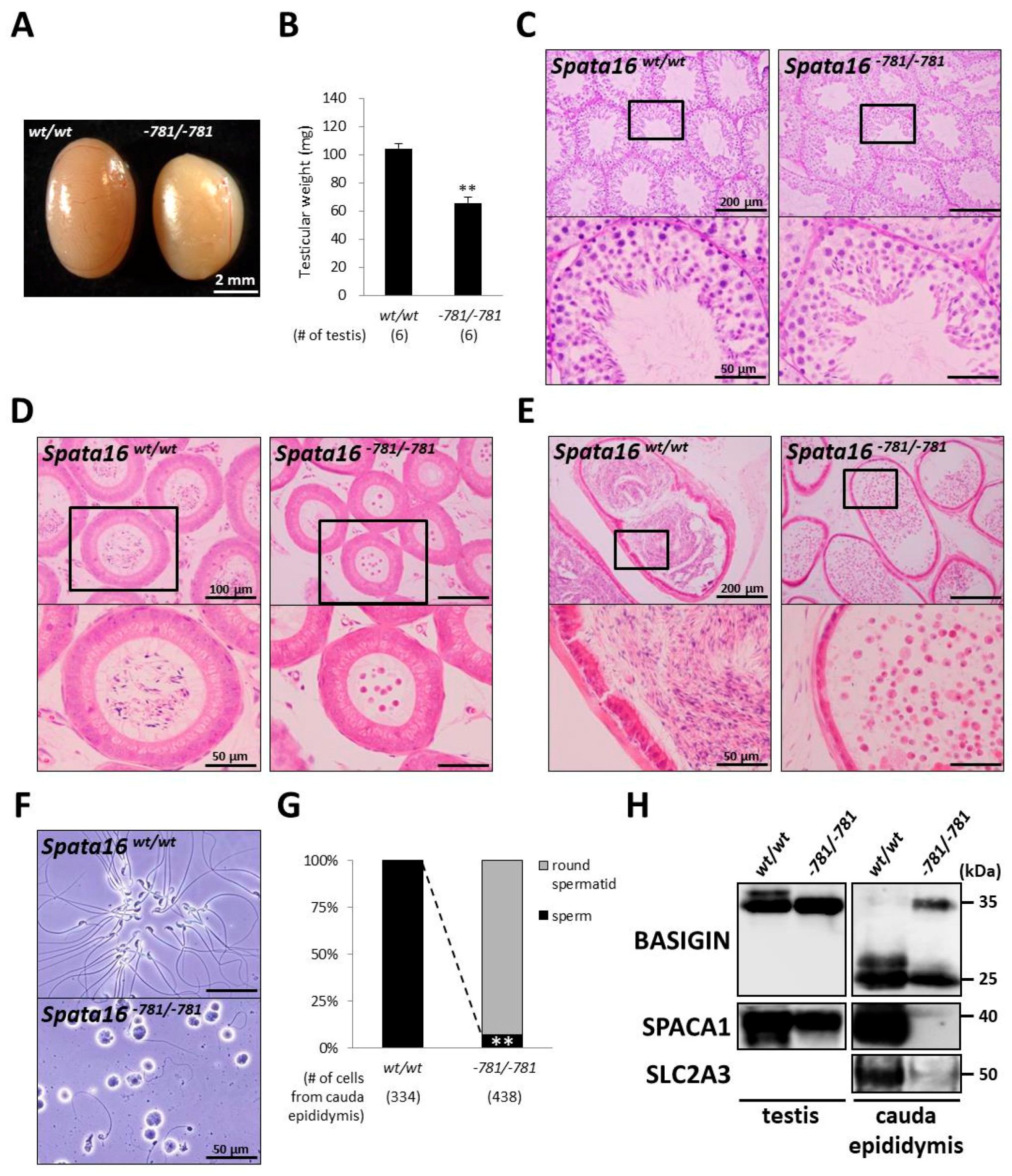
2.4. Spermiogenic Arrest in Spata16−781/−781 Mice
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. RT-PCR
4.3. Protein Sequence Alignment among Mammals
4.4. Construction of the HDR Donor Plasmid for Point Mutation in Spata16
4.5. Generation of Spata16 Mutant Mice with CRISPR/Cas9
4.6. Male Fertility Test
4.7. Testis Weights, Testis Histology, and Sperm Morphology
4.8. Antibodies
4.9. Immunostaining
4.10. Immunoblot
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Elinati, E.; Kuentz, P.; Redin, C.; Jaber, S.; Vanden Meerschaut, F.; Makarian, J.; Koscinski, I.; Nasr-Esfahani, M.H.; Demirol, A.; Gurgan, T.; et al. Globozoospermia is mainly due to DPY19L2 deletion via non-allelic homologous recombination involving two recombination hotspots. Hum. Mol. Genet. 2012, 21, 3695–3702. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Shi, Q.W.; Lu, G.X. A newly discovered mutation in PICK1 in a human with globozoospermia. Asian J. Androl. 2010, 12, 556–560. [Google Scholar] [CrossRef] [PubMed]
- Dam, A.H.; Koscinski, I.; Kremer, J.A.; Moutou, C.; Jaeger, A.S.; Oudakker, A.R.; Tournaye, H.; Charlet, N.; Lagier-Tourenne, C.; van Bokhoven, H.; et al. Homozygous mutation in SPATA16 is associated with male infertility in human globozoospermia. Am. J. Hum. Genet. 2007, 81, 813–820. [Google Scholar] [CrossRef] [PubMed]
- Coutton, C.; Escoffier, J.; Martinez, G.; Arnoult, C.; Ray, P.F. Teratozoospermia: Spotlight on the main genetic actors in the human. Hum. Reprod. Updat. 2015, 21, 455–485. [Google Scholar] [CrossRef] [PubMed]
- Dam, A.H.; Feenstra, I.; Westphal, J.R.; Ramos, L.; van Golde, R.J.; Kremer, J.A. Globozoospermia revisited. Hum. Reprod. Updat. 2007, 13, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Takeya, R.; Takeshige, K.; Sumimoto, H. Interaction of the PDZ domain of human PICK1 with class I ADP-ribosylation factors. Biochem. Biophys. Res. Commun. 2000, 267, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Staudinger, J.; Zhou, J.; Burgess, R.; Elledge, S.J.; Olson, E.N. PICK1: A perinuclear binding protein and substrate for protein kinase C isolated by the yeast two-hybrid system. J. Cell Biol. 1995, 128, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Zhang, N.; Wang, Y.N.; Shen, Y.; Wang, Y. Multiple faces of protein interacting with C kinase 1 (PICK1): Structure, function, and diseases. Neurochem. Int. 2016, 98, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, J.P.; Takamiya, K.; Shen, Y.; Xia, J.; Rubio, M.E.; Yu, S.; Jin, W.; Thomas, G.M.; Linden, D.J.; Huganir, R.L. Targeted in vivo mutations of the AMPA receptor subunit GluR2 and its interacting protein PICK1 eliminate cerebellar long-term depression. Neuron 2006, 49, 845–860. [Google Scholar] [CrossRef] [PubMed]
- Xiao, N.; Kam, C.; Shen, C.; Jin, W.; Wang, J.; Lee, K.M.; Jiang, L.; Xia, J. PICK1 deficiency causes male infertility in mice by disrupting acrosome formation. J. Clin. Investig. 2009, 119, 802–812. [Google Scholar] [CrossRef] [PubMed]
- Harbuz, R.; Zouari, R.; Pierre, V.; Ben Khelifa, M.; Kharouf, M.; Coutton, C.; Merdassi, G.; Abada, F.; Escoffier, J.; Nikas, Y.; et al. A recurrent deletion of DPY19L2 causes infertility in man by blocking sperm head elongation and acrosome formation. Am. J. Hum. Genet. 2011, 88, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Pierre, V.; Martinez, G.; Coutton, C.; Delaroche, J.; Yassine, S.; Novella, C.; Pernet-Gallay, K.; Hennebicq, S.; Ray, P.F.; Arnoult, C. Absence of Dpy19l2, a new inner nuclear membrane protein, causes globozoospermia in mice by preventing the anchoring of the acrosome to the nucleus. Development 2012, 139, 2955–2965. [Google Scholar] [CrossRef] [PubMed]
- Koscinski, I.; Elinati, E.; Fossard, C.; Redin, C.; Muller, J.; Velez de la Calle, J.; Schmitt, F.; Ben Khelifa, M.; Ray, P.F.; Kilani, Z.; et al. DPY19L2 deletion as a major cause of globozoospermia. Am. J. Hum. Genet. 2011, 88, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Ounis, L.; Zoghmar, A.; Coutton, C.; Rouabah, L.; Hachemi, M.; Martinez, D.; Martinez, G.; Bellil, I.; Khelifi, D.; Arnoult, C.; et al. Mutations of the aurora kinase C gene causing macrozoospermia are the most frequent genetic cause of male infertility in Algerian men. Asian J. Androl. 2015, 17, 68–73. [Google Scholar] [PubMed]
- Zhu, F.; Gong, F.; Lin, G.; Lu, G. DPY19L2 gene mutations are a major cause of globozoospermia: identification of three novel point mutations. Mol. Hum. Reprod. 2013, 19, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Noveski, P.; Madjunkova, S.; Maleva, I.; Sotiroska, V.; Petanovski, Z.; Plaseska-Karanfilska, D. A Homozygous Deletion of the DPY19l2 Gene is a Cause of Globozoospermia in Men from the Republic of Macedonia. Balk. J. Med. Genet. 2013, 16, 73–76. [Google Scholar] [CrossRef] [PubMed]
- Coutton, C.; Zouari, R.; Abada, F.; Ben Khelifa, M.; Merdassi, G.; Triki, C.; Escalier, D.; Hesters, L.; Mitchell, V.; Levy, R.; et al. MLPA and sequence analysis of DPY19L2 reveals point mutations causing globozoospermia. Hum. Reprod. 2012, 27, 2549–2558. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Xiao, J.; Chen, J.; Li, J.; Yin, L.; Zhu, H.; Zhou, Z.; Sha, J. Identification and characterization of a novel human testis-specific Golgi protein, NYD-SP12. Mol. Hum. Reprod. 2003, 9, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Lin, M.; Xu, M.; Zhou, Z.M.; Sha, J.H. Gene functional research using polyethylenimine-mediated in vivo gene transfection into mouse spermatogenic cells. Asian J. Androl. 2006, 8, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wan, H.; Li, X.; Liu, W.; Chen, Q.; Wang, Y.; Yang, L.; Tang, H.; Zhang, X.; Duan, E.; et al. Atg7 is required for acrosome biogenesis during spermatogenesis in mice. Cell Res. 2014, 24, 852–869. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Toselli, P.A.; Russell, L.D.; Seldin, D.C. Globozoospermia in mice lacking the casein kinase II alpha’ catalytic subunit. Nat. Genet. 1999, 23, 118–121. [Google Scholar] [PubMed]
- Yildiz, Y.; Matern, H.; Thompson, B.; Allegood, J.C.; Warren, R.L.; Ramirez, D.M.; Hammer, R.E.; Hamra, F.K.; Matern, S.; Russell, D.W. Mutation of beta-glucosidase 2 causes glycolipid storage disease and impaired male fertility. J. Clin. Investig. 2006, 116, 2985–2994. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Liu, C.; Zhang, L.; Chen, M.; Zhou, Y.; Qin, Y.; Wang, Y.; Duo, S.; Cui, X.; Bao, S.; et al. Globozoospermia and lack of acrosome formation in GM130-deficient mice. Cell Death Dis. 2017, 8, e2532. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.; Ito, C.; Natsume, Y.; Sugitani, Y.; Yamanaka, H.; Kuretake, S.; Yanagida, K.; Sato, A.; Toshimori, K.; Noda, T. Lack of acrosome formation in mice lacking a Golgi protein, GOPC. Proc. Natl. Acad. Sci. USA 2002, 99, 11211–11216. [Google Scholar] [CrossRef] [PubMed]
- Kang-Decker, N.; Mantchev, G.T.; Juneja, S.C.; McNiven, M.A.; van Deursen, J.M. Lack of acrosome formation in Hrb-deficient mice. Science 2001, 294, 1531–1533. [Google Scholar] [CrossRef] [PubMed]
- Audouard, C.; Christians, E. Hsp90beta1 knockout targeted to male germline: A mouse model for globozoospermia. Fertil. Steril. 2011, 95, 1475–1477.e4. [Google Scholar] [CrossRef] [PubMed]
- Doran, J.; Walters, C.; Kyle, V.; Wooding, P.; Hammett-Burke, R.; Colledge, W.H. Mfsd14a (Hiat1) gene disruption causes globozoospermia and infertility in male mice. Reproduction 2016, 152, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Song, Z.; Wang, L.; Yu, H.; Liu, W.; Shang, Y.; Xu, Z.; Zhao, H.; Gao, F.; Wen, J.; et al. Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 2017, 144, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Oberheide, K.; Puchkov, D.; Jentsch, T.J. Loss of the Na+/H+ exchanger NHE8 causes male infertility in mice by disrupting acrosome formation. J. Biol. Chem. 2017, 292, 10845–10854. [Google Scholar] [CrossRef] [PubMed]
- Funaki, T.; Kon, S.; Tanabe, K.; Natsume, W.; Sato, S.; Shimizu, T.; Yoshida, N.; Wong, W.F.; Ogura, A.; Ogawa, T.; et al. The Arf GAP SMAP2 is necessary for organized vesicle budding from the trans-Golgi network and subsequent acrosome formation in spermiogenesis. Mol. Biol. Cell 2013, 24, 2633–2644. [Google Scholar] [CrossRef] [PubMed]
- Fujihara, Y.; Satouh, Y.; Inoue, N.; Isotani, A.; Ikawa, M.; Okabe, M. SPACA1-deficient male mice are infertile with abnormally shaped sperm heads reminiscent of globozoospermia. Development 2012, 139, 3583–3589. [Google Scholar] [CrossRef] [PubMed]
- Lerer-Goldshtein, T.; Bel, S.; Shpungin, S.; Pery, E.; Motro, B.; Goldstein, R.S.; Bar-Sheshet, S.I.; Breitbart, H.; Nir, U. TMF/ARA160: A key regulator of sperm development. Dev. Biol. 2010, 348, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Paiardi, C.; Pasini, M.E.; Gioria, M.; Berruti, G. Failure of acrosome formation and globozoospermia in the wobbler mouse, a Vps54 spontaneous recessive mutant. Spermatogenesis 2011, 1, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.N.; Roy, A.; Yan, W.; Burns, K.H.; Matzuk, M.M. Loss of zona pellucida binding proteins in the acrosomal matrix disrupts acrosome biogenesis and sperm morphogenesis. Mol. Cell. Biol. 2007, 27, 6794–6805. [Google Scholar] [CrossRef] [PubMed]
- Yatsenko, A.N.; O'Neil, D.S.; Roy, A.; Arias-Mendoza, P.A.; Chen, R.; Murthy, L.J.; Lamb, D.J.; Matzuk, M.M. Association of mutations in the zona pellucida binding protein 1 (ZPBP1) gene with abnormal sperm head morphology in infertile men. Mol. Hum. Reprod. 2012, 18, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Cesario, M.M.; Bartles, J.R. Compartmentalization, processing and redistribution of the plasma membrane protein CE9 on rodent spermatozoa. Relationship of the annulus to domain boundaries in the plasma membrane of the tail. J. Cell Sci. 1994, 107 Pt 2, 561–570. [Google Scholar] [PubMed]
- Miyata, H.; Castaneda, J.M.; Fujihara, Y.; Yu, Z.; Archambeault, D.R.; Isotani, A.; Kiyozumi, D.; Kriseman, M.L.; Mashiko, D.; Matsumura, T.; et al. Genome engineering uncovers 54 evolutionarily conserved and testis-enriched genes that are not required for male fertility in mice. Proc. Natl. Acad. Sci. USA 2016, 113, 7704–7710. [Google Scholar] [CrossRef] [PubMed]
- Oji, A.; Noda, T.; Fujihara, Y.; Miyata, H.; Kim, Y.J.; Muto, M.; Nozawa, K.; Matsumura, T.; Isotani, A.; Ikawa, M. CRISPR/Cas9 mediated genome editing in ES cells and its application for chimeric analysis in mice. Sci. Rep. 2016, 6, 31666. [Google Scholar] [CrossRef] [PubMed]
- Fujihara, Y.; Kaseda, K.; Inoue, N.; Ikawa, M.; Okabe, M. Production of mouse pups from germline transmission-failed knockout chimeras. Transgenic Res. 2013, 22, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Naito, Y.; Hino, K.; Bono, H.; Ui-Tei, K. CRISPRdirect: Software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 2015, 31, 1120–1123. [Google Scholar] [CrossRef] [PubMed]
- Fujihara, Y.; Tokuhiro, K.; Muro, Y.; Kondoh, G.; Araki, Y.; Ikawa, M.; Okabe, M. Expression of TEX101, regulated by ACE, is essential for the production of fertile mouse spermatozoa. Proc. Natl. Acad. Sci. USA 2013, 110, 8111–8116. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, R.; Yamagata, K.; Hasuwa, H.; Inano, E.; Ikawa, M.; Okabe, M. Cd52, known as a major maturation-associated sperm membrane antigen secreted from the epididymis, is not required for fertilization in the mouse. Genes Cells 2008, 13, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Muto, M.; Fujihara, Y.; Tobita, T.; Kiyozumi, D.; Ikawa, M. Lentiviral Vector-mediated complementation restored fetal viability but not placental hyperplasia in Plac1-deficient mice. Biol. Reprod. 2016, 94, 6. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, R.; Yamagata, K.; Ikawa, M.; Moss, S.B.; Okabe, M. Aberrant distribution of ADAM3 in sperm from both angiotensin-converting enzyme (Ace)- and calmegin (Clgn)-deficient mice. Biol. Reprod. 2006, 75, 760–766. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fujihara, Y.; Oji, A.; Larasati, T.; Kojima-Kita, K.; Ikawa, M. Human Globozoospermia-Related Gene Spata16 Is Required for Sperm Formation Revealed by CRISPR/Cas9-Mediated Mouse Models. Int. J. Mol. Sci. 2017, 18, 2208. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18102208
Fujihara Y, Oji A, Larasati T, Kojima-Kita K, Ikawa M. Human Globozoospermia-Related Gene Spata16 Is Required for Sperm Formation Revealed by CRISPR/Cas9-Mediated Mouse Models. International Journal of Molecular Sciences. 2017; 18(10):2208. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18102208
Chicago/Turabian StyleFujihara, Yoshitaka, Asami Oji, Tamara Larasati, Kanako Kojima-Kita, and Masahito Ikawa. 2017. "Human Globozoospermia-Related Gene Spata16 Is Required for Sperm Formation Revealed by CRISPR/Cas9-Mediated Mouse Models" International Journal of Molecular Sciences 18, no. 10: 2208. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18102208